 |  |
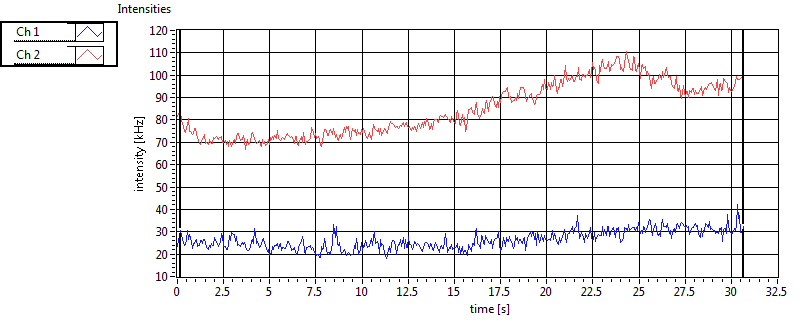
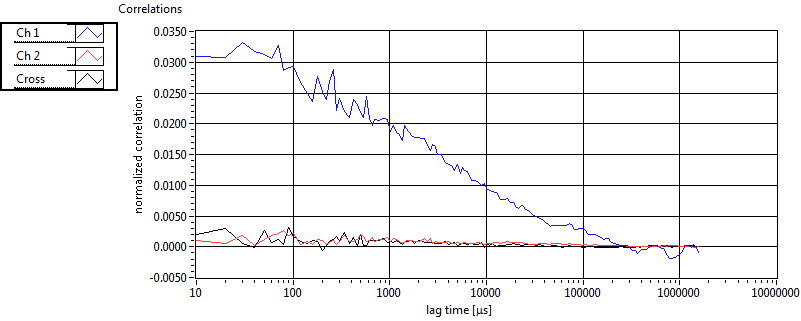
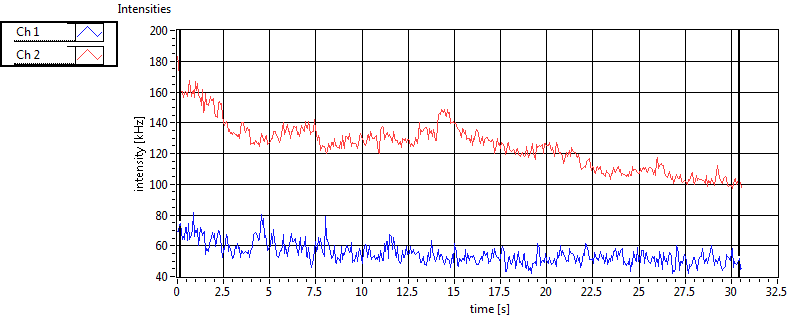
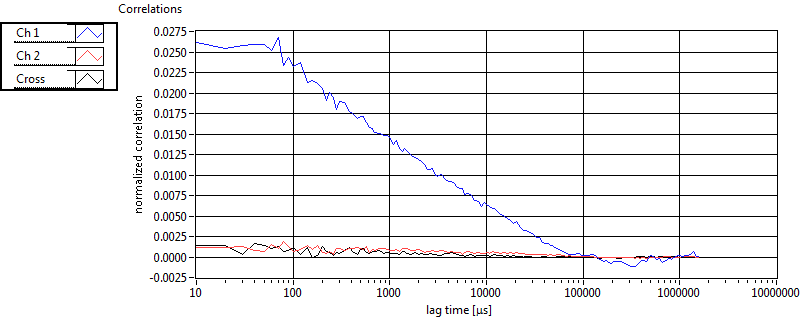
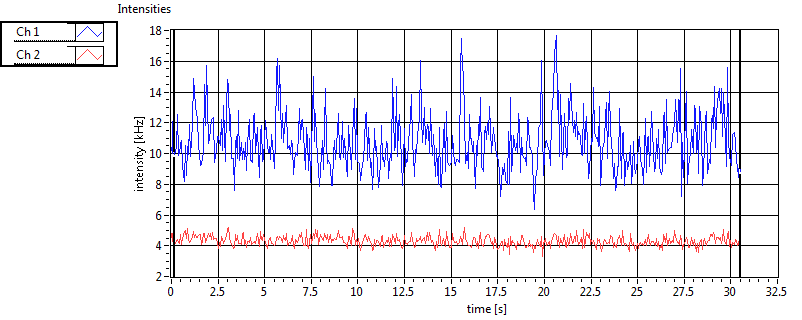
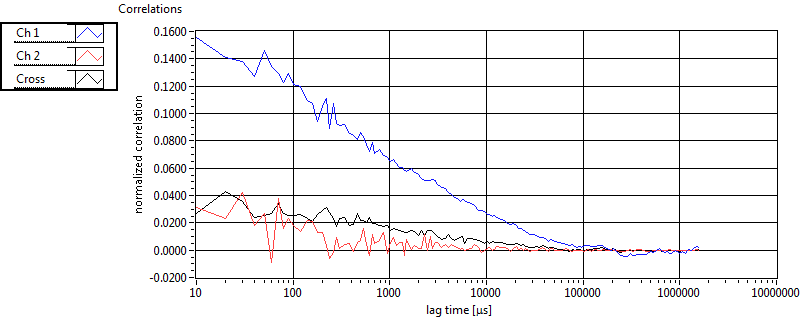
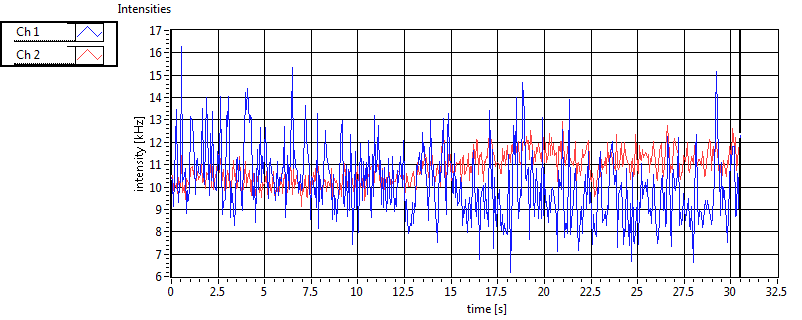
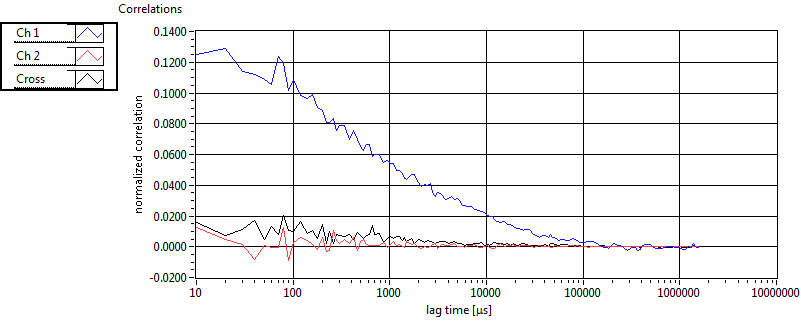
| 1 | 20110915\U03--V00--0827 |
|---|---|
| 2 | 20110920\U03--V01--0827 |
| 3 | 20110921\U02--V00--0827 |
| 4 | 20110927\U00--V01--0827 |
| Ch1 ave Nuc |
Ch1 sd Nuc |
Ch2 ave Nuc |
Ch2 sd Nuc |
Cross ave Nuc |
Cross sd Nuc |
Ch1 ave Cyto |
Ch1 sd Cyto |
Ch2 ave Cyto |
Ch2 sd Cyto |
Cross ave Cyto |
Cross sd Cyto |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 1.348E+2 | 9.322E+1 | 9.527E+2 | 4.228E+2 | 2.301E+1 | 2.039E+1 | 2.528E+1 | 3.346E+1 | 3.705E+2 | 2.185E+2 | 1.271E+1 | 1.635E+1 |
| N_corr | 1.766E+2 | 1.377E+2 | 1.291E+3 | 7.119E+2 | 3.867E+1 | 3.768E+1 | 3.292E+1 | 5.450E+1 | 5.194E+2 | 2.960E+2 | 1.640E+1 | 2.060E+1 |
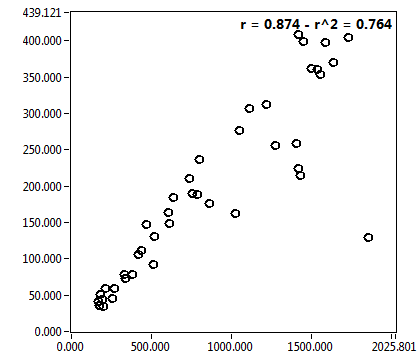
| c [nM] | 8.382E+2 | 5.797E+2 | 4.419E+3 | 1.961E+3 | 1.183E+2 | 1.048E+2 | 1.572E+2 | 2.081E+2 | 1.719E+3 | 1.013E+3 | 6.534E+1 | 8.405E+1 |
| c_corr [nM] | 1.098E+3 | 8.563E+2 | 5.988E+3 | 3.302E+3 | 1.988E+2 | 1.937E+2 | 2.048E+2 | 3.390E+2 | 2.409E+3 | 1.373E+3 | 8.431E+1 | 1.059E+2 |
| ratioG | 1.262E-1 | 6.501E-2 | 1.176E-1 | 7.297E-2 | ||||||||
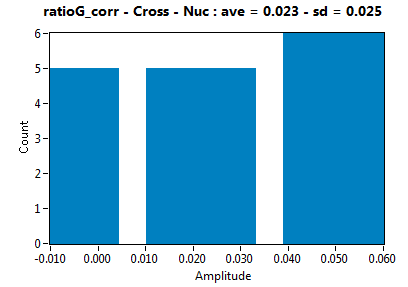
| ratioG_corr | 2.264E-2 | 2.543E-2 | 8.639E-2 | 5.540E-2 | ||||||||
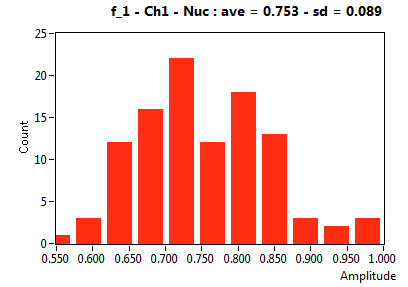
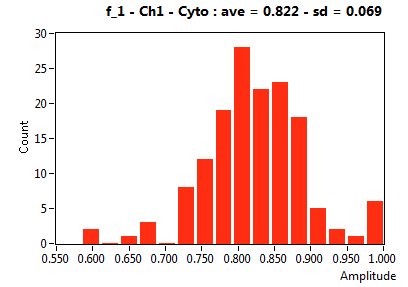
| f_1 | 7.530E-1 | 8.860E-2 | 5.414E-1 | 2.445E-1 | 9.535E-1 | 8.002E-2 | 8.218E-1 | 6.859E-2 | 3.699E-1 | 3.761E-1 | 8.703E-1 | 1.835E-1 |
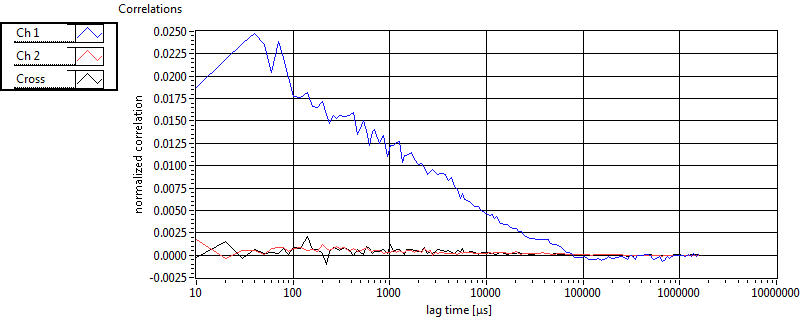
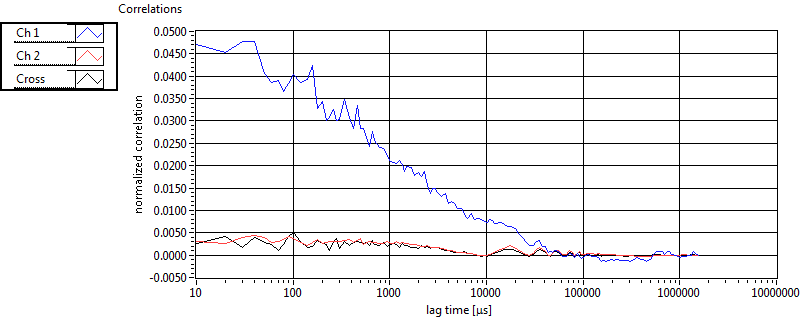
| tauD_1 [ms] | 1.969E+0 | 7.664E-1 | 4.686E+0 | 1.037E+1 | 1.277E+0 | 8.586E-1 | 1.086E+0 | 4.853E-1 | 1.800E+0 | 0.000E+0 | 2.364E+0 | 3.887E-1 |
| D_1 [mum^2/s] | 6.109E+0 | 2.055E+0 | 6.615E+0 | 1.839E+0 | 1.924E+1 | 2.687E+1 | 1.200E+1 | 8.907E+0 | 7.157E+0 | 0.000E+0 | 5.199E+0 | 7.808E-1 |
| alpha_1 | 9.560E-1 | 9.784E-2 | 8.940E-1 | 1.165E-1 | 7.924E-1 | 2.401E-1 | 9.239E-1 | 8.803E-2 | 9.200E-1 | 1.110E-16 | 1.200E+0 | 0.000E+0 |
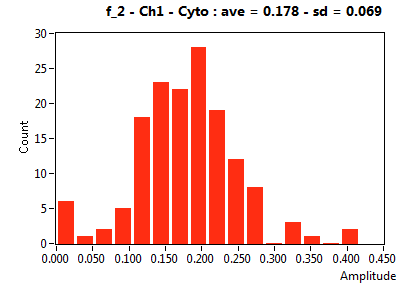
| f_2 | 2.470E-1 | 8.860E-2 | 4.586E-1 | 2.445E-1 | 4.646E-2 | 8.002E-2 | 1.782E-1 | 6.859E-2 | 6.301E-1 | 3.761E-1 | 1.297E-1 | 1.835E-1 |
| tauD_2 [ms] | 3.769E+1 | 2.217E+1 | 6.512E+1 | 5.769E+1 | 6.824E+1 | 5.464E+1 | 2.987E+1 | 1.846E+1 | 8.895E+1 | 9.386E+1 | 1.059E+2 | 1.005E+2 |
| D_2 [mum^2/s] | 3.389E-1 | 1.244E-1 | 7.719E-1 | 1.070E+0 | 2.650E-1 | 1.383E-1 | 4.318E-1 | 1.642E-1 | 1.844E+0 | 2.075E+0 | 2.516E-1 | 1.520E-1 |
| alpha_2 | 1.574E+0 | 3.177E-1 | 1.350E+0 | 3.675E-1 | 1.173E+0 | 2.218E-1 | 1.284E+0 | 2.347E-1 | 1.710E+0 | 3.721E-1 | 1.239E+0 | 2.424E-1 |
| tauD_m [ms] | 1.036E+1 | 5.561E+0 | 3.627E+1 | 3.706E+1 | 4.465E+0 | 8.115E+0 | 6.374E+0 | 5.130E+0 | 5.589E+1 | 6.804E+1 | 3.425E+1 | 4.485E+1 |
| D_m [mum^2/s] | 4.625E+0 | 1.505E+0 | 3.702E+0 | 1.556E+0 | 1.833E+1 | 2.678E+1 | 9.728E+0 | 5.591E+0 | 4.148E+0 | 2.324E+0 | 4.432E+0 | 7.858E-1 |
| R [1/s] | 1.198E+1 | 1.295E+1 | 9.581E+0 | 6.816E+0 | ||||||||
| CPM [kHz] | 1.278E+0 | 3.662E-1 | 3.126E-1 | 2.539E-1 | 1.045E+0 | 4.198E-1 | 2.959E-1 | 2.168E-1 | ||||
| CPM_corr [kHz] | 1.302E+0 | 3.675E-1 | 3.584E-1 | 2.681E-1 | 1.205E+0 | 4.645E-1 | 3.368E-1 | 2.205E-1 | ||||
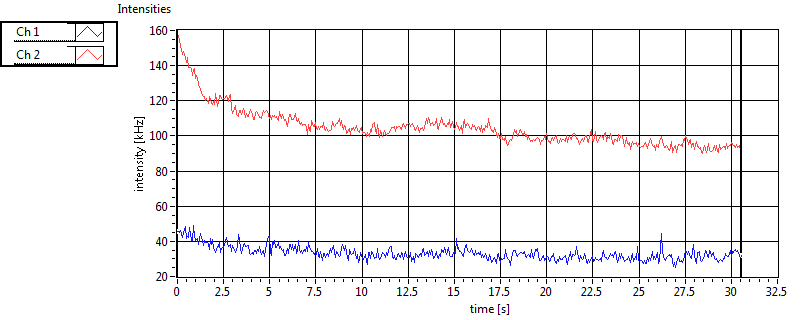
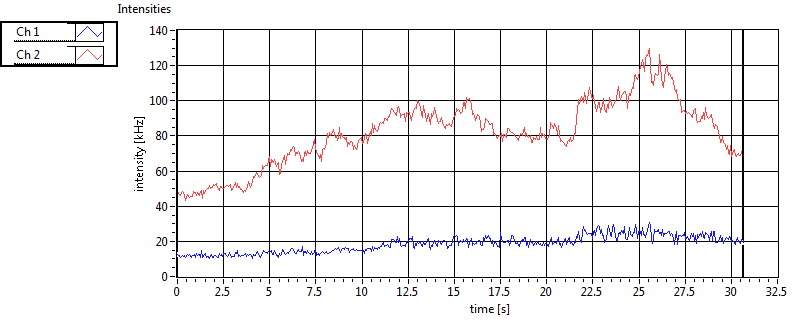
| Int [kHz] | 1.815E+2 | 1.289E+2 | 1.517E+2 | 3.183E+1 | 2.630E+1 | 4.146E+1 | 7.174E+1 | 5.086E+1 | ||||
| Int_corr [kHz] | 2.355E+2 | 1.791E+2 | 2.363E+2 | 6.755E+1 | 3.478E+1 | 5.426E+1 | 1.054E+2 | 5.697E+1 | ||||
| # samples | 105 | 105 | 25 | 25 | 16 | 16 | 150 | 150 | 5 | 5 | 3 | 3 |
| # cells | 54 |
![c_corr [nM] - Ch1 - Nuc : ave = 1098.142 - sd = 856.311](0827_c_corr_1n.png)
![Int_corr [kHz] - Ch1 - Nuc : ave = 235.536 - sd = 179.091](0827_Int_corr_1n.png)
![D_1 [mum^2/s] - Ch1 - Nuc : ave = 6.109 - sd = 2.055](0827_D_1_1n.png)
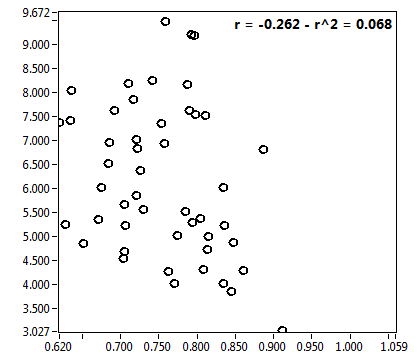
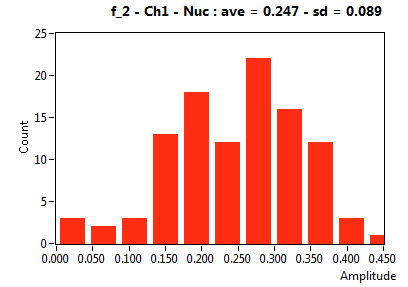
![D_2 [mum^2/s] - Ch1 - Nuc : ave = 0.339 - sd = 0.124](0827_D_2_1n.png)
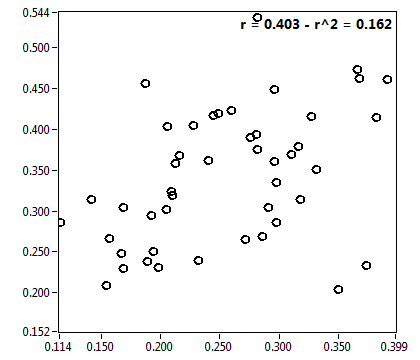
![CPM_corr [kHz] - Ch1 - Nuc : ave = 1.302 - sd = 0.368](0827_CPM_corr_1n.png)
![D_m [mum^2/s] - Ch1 - Nuc : ave = 4.625 - sd = 1.505](0827_D_m_1n.png)
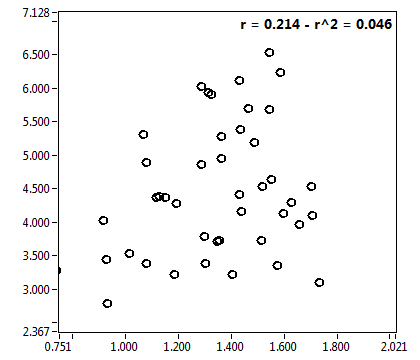
![c_corr [nM] - Ch1 - Nuc : ave = 1098.142 - sd = 856.311](0827_c_corr_1n.png)

![c_corr [nM] - Ch1 - Cyto : ave = 204.758 - sd = 338.971](0827_c_corr_1c.png)
![Int_corr [kHz] - Ch1 - Cyto : ave = 34.782 - sd = 54.261](0827_Int_corr_1c.png)
![D_1 [mum^2/s] - Ch1 - Cyto : ave = 11.999 - sd = 8.907](0827_D_1_1c.png)
![D_2 [mum^2/s] - Ch1 - Cyto : ave = 0.432 - sd = 0.164](0827_D_2_1c.png)
![CPM_corr [kHz] - Ch1 - Cyto : ave = 1.205 - sd = 0.464](0827_CPM_corr_1c.png)
![D_m [mum^2/s] - Ch1 - Cyto : ave = 9.728 - sd = 5.591](0827_D_m_1c.png)
