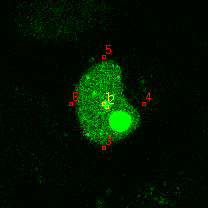
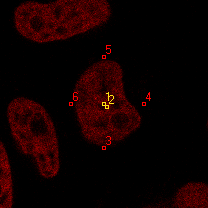
 |  |
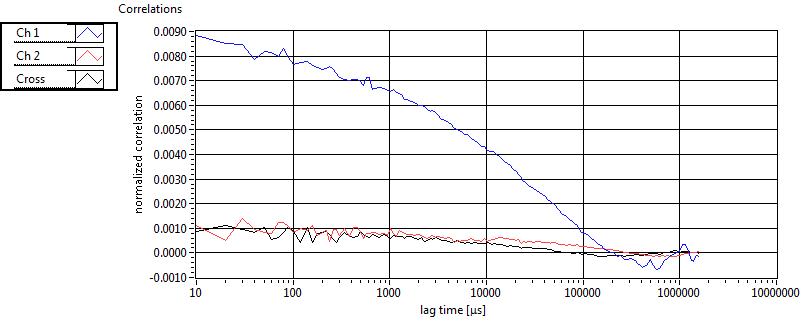
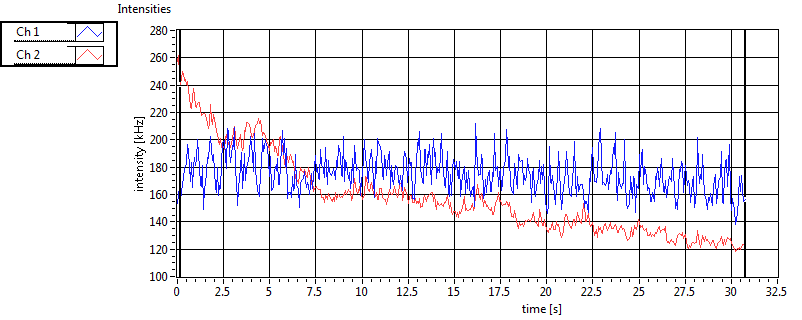
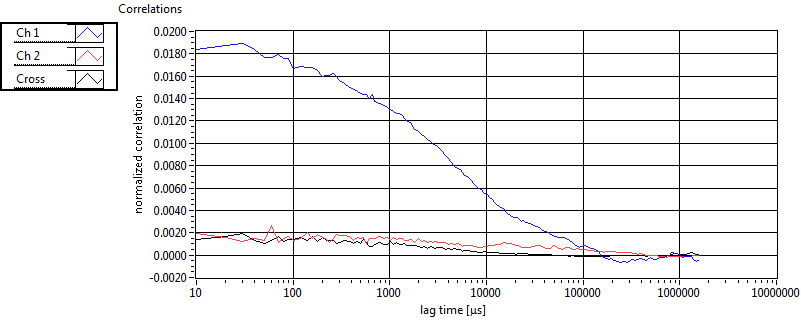
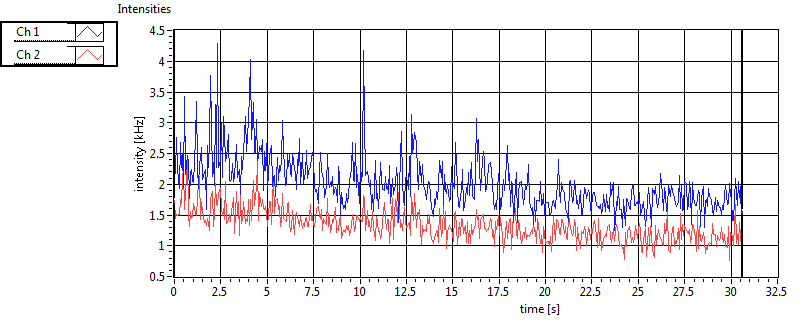
| 1 | 20110914\U01--V00--1499 |
|---|---|
| 2 | 20111111\U03--V00--1499 |
| 3 | 20111115\U03--V00--1499 |
| 4 | 20111116\U03--V00--1499 |
| 5 | 20111130\U01--V00--1499 |
| 6 | 20111207\U00--V01--1499 |
| 7 | 20111213\U02--V00--1499 |
| 8 | 20120131\U02--V00--1499 |
| 9 | 20120222\U02--V00--1499 |
| 10 | 20120229\U01--V00--1499 |
| 11 | 20120309\U01--V00--1499 |
| 12 | 20120322\U01--V00--1499 |
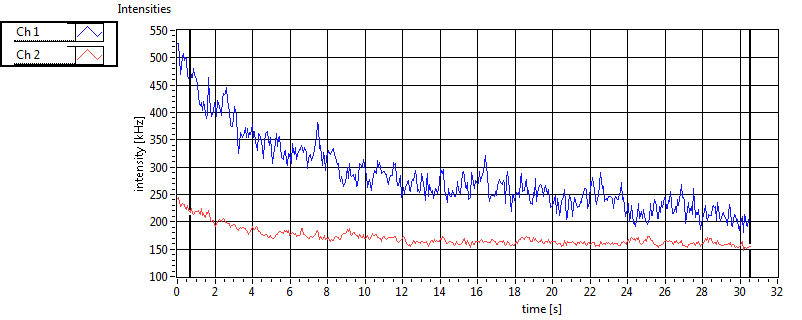
| Ch1 ave Nuc |
Ch1 sd Nuc |
Ch2 ave Nuc |
Ch2 sd Nuc |
Cross ave Nuc |
Cross sd Nuc |
Ch1 ave Cyto |
Ch1 sd Cyto |
Ch2 ave Cyto |
Ch2 sd Cyto |
Cross ave Cyto |
Cross sd Cyto |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 1.429E+2 | 1.924E+2 | 1.099E+3 | 6.566E+2 | 2.535E+1 | 1.329E+2 | 2.760E+1 | 4.495E+1 | 2.843E+2 | 2.646E+2 | 5.959E+0 | 4.861E+0 |
| N_corr | 2.362E+2 | 4.045E+2 | 1.625E+3 | 1.078E+3 | 6.558E+1 | 3.154E+2 | 3.165E+1 | 6.051E+1 | 3.837E+2 | 4.124E+2 | 9.376E+0 | 9.486E+0 |
| c [nM] | 8.890E+2 | 1.196E+3 | 5.096E+3 | 3.045E+3 | 1.303E+2 | 6.831E+2 | 1.717E+2 | 2.796E+2 | 1.319E+3 | 1.227E+3 | 3.063E+1 | 2.499E+1 |
| c_corr [nM] | 1.469E+3 | 2.516E+3 | 7.539E+3 | 5.000E+3 | 3.371E+2 | 1.622E+3 | 1.968E+2 | 3.763E+2 | 1.780E+3 | 1.913E+3 | 4.820E+1 | 4.877E+1 |
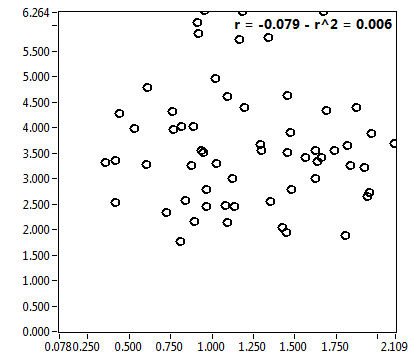
| ratioG | 1.016E-1 | 1.353E-1 | 2.542E-1 | 2.008E-1 | ||||||||
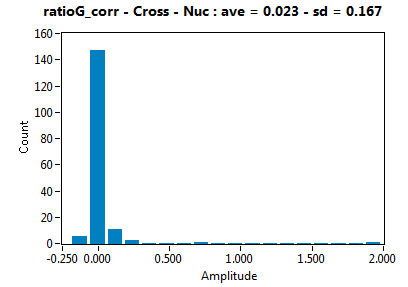
| ratioG_corr | 2.261E-2 | 1.670E-1 | 6.808E-2 | 5.696E-2 | ||||||||
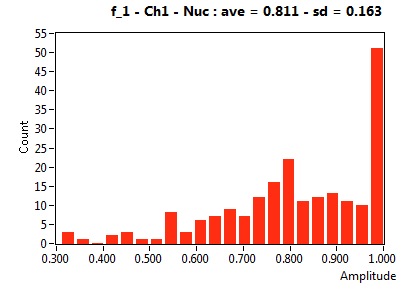
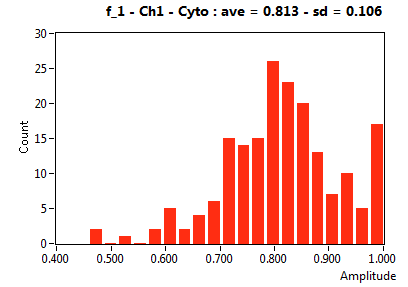
| f_1 | 8.106E-1 | 1.630E-1 | 4.004E-1 | 2.137E-1 | 9.456E-1 | 9.527E-2 | 8.126E-1 | 1.058E-1 | 7.264E-1 | 2.744E-1 | 8.561E-1 | 9.379E-2 |
| tauD_1 [ms] | 3.952E+0 | 1.737E+0 | 1.838E+0 | 2.890E-2 | 3.365E+0 | 2.656E+0 | 2.708E+0 | 1.835E+0 | 6.412E+0 | 1.250E+1 | 2.416E+0 | 1.299E+0 |
| D_1 [mum^2/s] | 3.369E+0 | 2.245E+0 | 7.010E+0 | 1.112E-1 | 6.192E+0 | 7.705E+0 | 7.069E+0 | 1.170E+1 | 5.842E+0 | 2.261E+0 | 1.161E+1 | 2.057E+1 |
| alpha_1 | 9.921E-1 | 1.027E-1 | 9.200E-1 | 2.596E-4 | 8.760E-1 | 1.920E-1 | 9.045E-1 | 1.128E-1 | 9.009E-1 | 1.453E-1 | 8.527E-1 | 2.298E-1 |
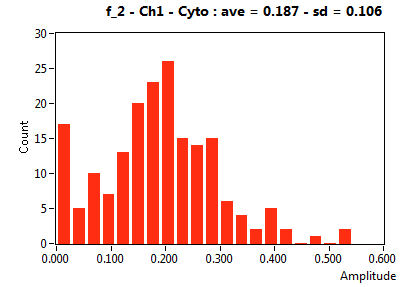
| f_2 | 1.894E-1 | 1.630E-1 | 5.996E-1 | 2.137E-1 | 5.443E-2 | 9.527E-2 | 1.874E-1 | 1.058E-1 | 2.736E-1 | 2.744E-1 | 1.439E-1 | 9.379E-2 |
| tauD_2 [ms] | 4.337E+1 | 2.836E+1 | 9.173E+1 | 1.085E+2 | 9.528E+1 | 5.251E+1 | 4.547E+1 | 3.157E+1 | 7.377E+1 | 1.038E+2 | 7.049E+1 | 4.623E+1 |
| D_2 [mum^2/s] | 3.549E-1 | 2.984E-1 | 5.019E-1 | 7.375E-1 | 1.683E-1 | 1.310E-1 | 3.156E-1 | 1.731E-1 | 9.031E-1 | 1.017E+0 | 2.297E-1 | 1.198E-1 |
| alpha_2 | 1.467E+0 | 3.249E-1 | 1.286E+0 | 3.206E-1 | 1.236E+0 | 2.962E-1 | 1.387E+0 | 2.996E-1 | 1.236E+0 | 3.488E-1 | 1.657E+0 | 3.911E-1 |
| tauD_m [ms] | 1.102E+1 | 9.402E+0 | 5.493E+1 | 7.297E+1 | 9.309E+0 | 1.600E+1 | 1.065E+1 | 8.896E+0 | 3.273E+1 | 3.876E+1 | 1.372E+1 | 1.574E+1 |
| D_m [mum^2/s] | 2.631E+0 | 1.174E+0 | 3.159E+0 | 1.322E+0 | 5.902E+0 | 7.595E+0 | 5.632E+0 | 8.457E+0 | 4.031E+0 | 2.088E+0 | 1.049E+1 | 2.004E+1 |
| R [1/s] | 1.834E+1 | 3.438E+1 | 4.588E+0 | 6.967E+0 | ||||||||
| CPM [kHz] | 2.336E+0 | 9.262E-1 | 3.928E-1 | 3.540E-1 | 9.507E-1 | 7.153E-1 | 2.865E-1 | 2.102E-1 | ||||
| CPM_corr [kHz] | 2.366E+0 | 9.311E-1 | 4.622E-1 | 4.135E-1 | 1.215E+0 | 7.584E-1 | 5.475E-1 | 5.096E-1 | ||||
| Int [kHz] | 2.296E+2 | 1.727E+2 | 2.111E+2 | 6.790E+1 | 1.932E+1 | 2.526E+1 | 6.937E+1 | 6.455E+1 | ||||
| Int_corr [kHz] | 3.481E+2 | 4.078E+2 | 3.628E+2 | 1.355E+2 | 2.423E+1 | 3.071E+1 | 1.117E+2 | 1.151E+2 | ||||
| # samples | 209 | 209 | 175 | 175 | 167 | 167 | 187 | 187 | 31 | 31 | 22 | 22 |
| # cells | 127 |
![c_corr [nM] - Ch1 - Nuc : ave = 1469.032 - sd = 2515.980](1499_c_corr_1n.png)
![Int_corr [kHz] - Ch1 - Nuc : ave = 348.108 - sd = 407.762](1499_Int_corr_1n.png)

![D_1 [mum^2/s] - Ch1 - Nuc : ave = 3.369 - sd = 2.245](1499_D_1_1n.png)
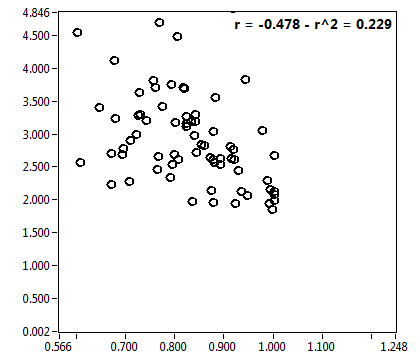
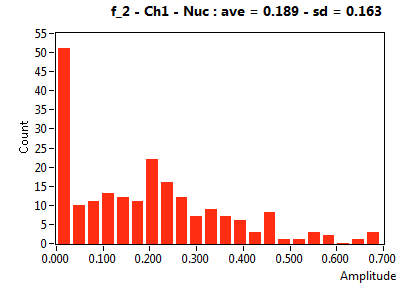
![D_2 [mum^2/s] - Ch1 - Nuc : ave = 0.355 - sd = 0.298](1499_D_2_1n.png)
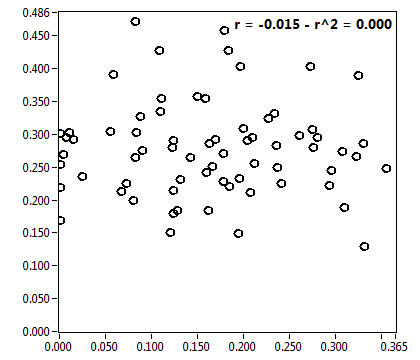
![CPM_corr [kHz] - Ch1 - Nuc : ave = 2.366 - sd = 0.931](1499_CPM_corr_1n.png)
![D_m [mum^2/s] - Ch1 - Nuc : ave = 2.631 - sd = 1.174](1499_D_m_1n.png)
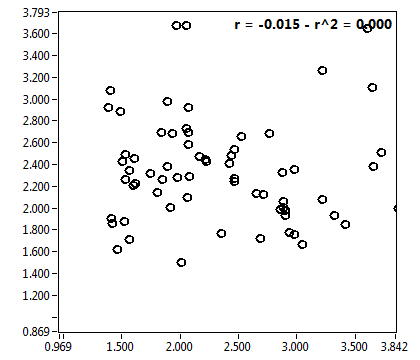
![c_corr [nM] - Ch1 - Nuc : ave = 1469.032 - sd = 2515.980](1499_c_corr_1n.png)
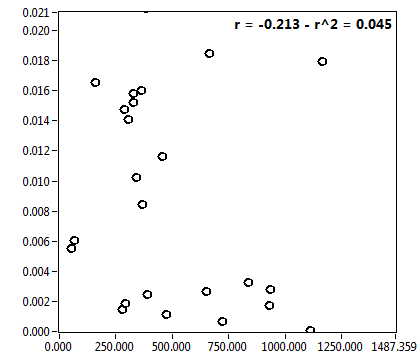
![c_corr [nM] - Ch1 - Cyto : ave = 196.810 - sd = 376.346](1499_c_corr_1c.png)
![Int_corr [kHz] - Ch1 - Cyto : ave = 24.229 - sd = 30.711](1499_Int_corr_1c.png)
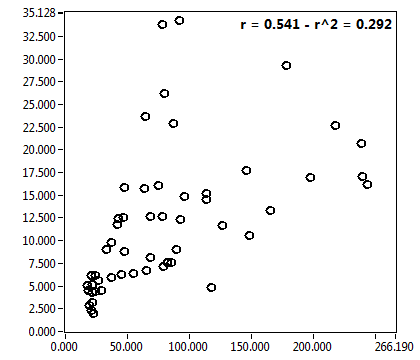
![D_1 [mum^2/s] - Ch1 - Cyto : ave = 7.069 - sd = 11.697](1499_D_1_1c.png)
![D_2 [mum^2/s] - Ch1 - Cyto : ave = 0.316 - sd = 0.173](1499_D_2_1c.png)
![CPM_corr [kHz] - Ch1 - Cyto : ave = 1.215 - sd = 0.758](1499_CPM_corr_1c.png)
![D_m [mum^2/s] - Ch1 - Cyto : ave = 5.632 - sd = 8.457](1499_D_m_1c.png)