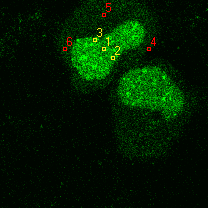
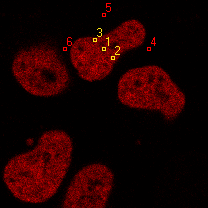
 |  |
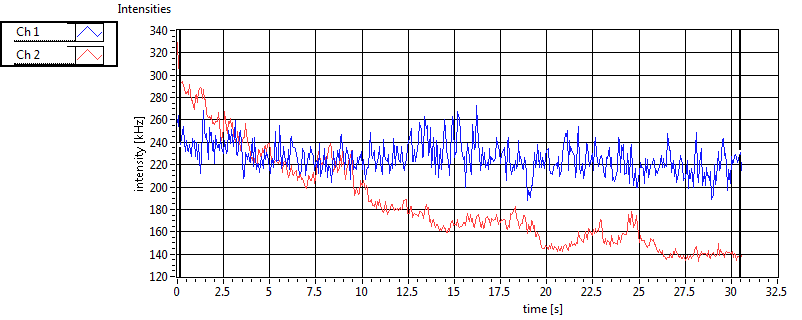
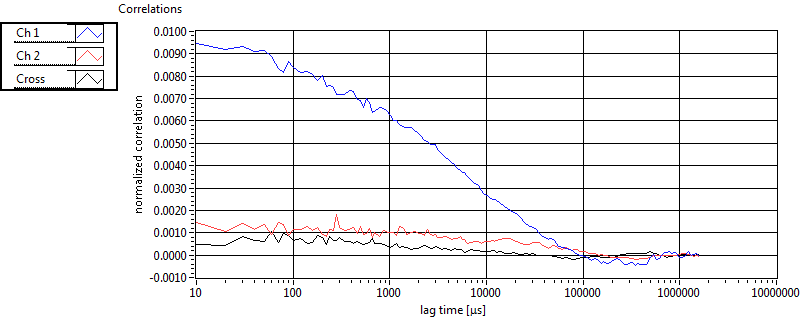
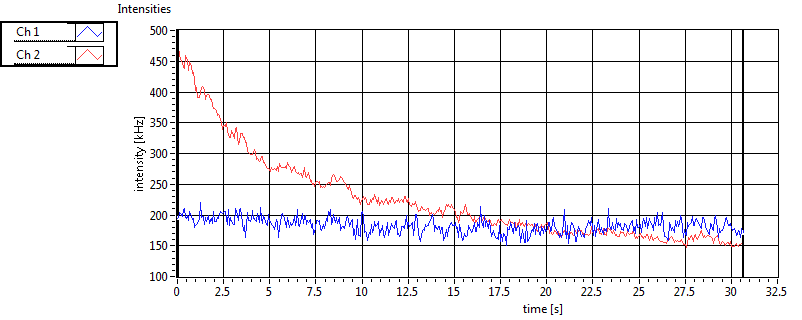
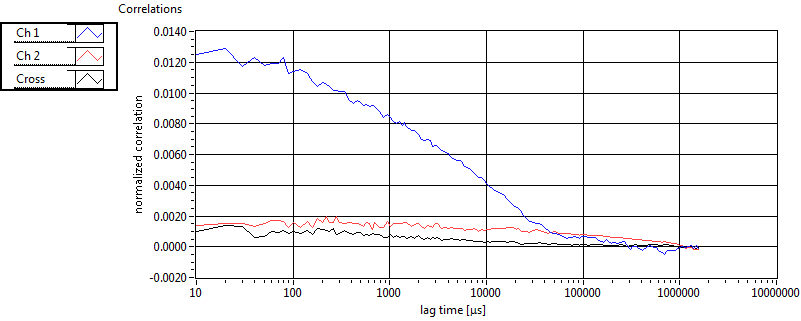
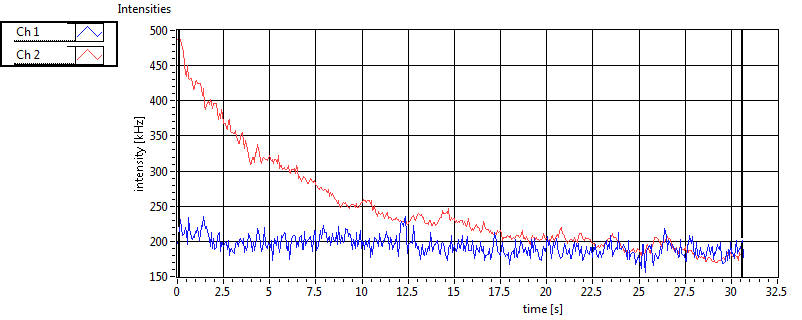
| 1 | 20111111\U01--V01--1909 |
|---|---|
| 2 | 20111115\U01--V01--1909 |
| 3 | 20111116\U01--V01--1909 |
| 4 | 20111215\U01--V00--1909 |
| Ch1 ave Nuc |
Ch1 sd Nuc |
Ch2 ave Nuc |
Ch2 sd Nuc |
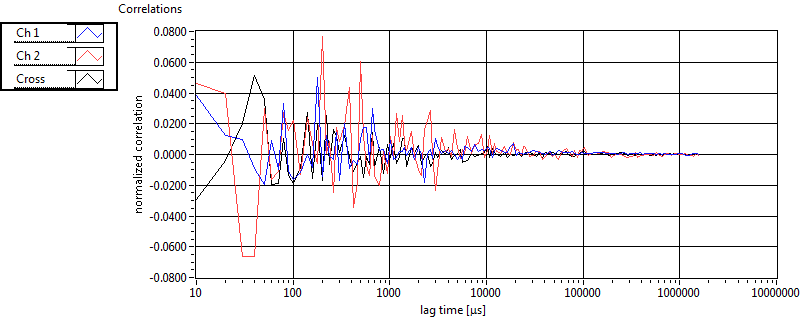
Cross ave Nuc |
Cross sd Nuc |
Ch1 ave Cyto |
Ch1 sd Cyto |
Ch2 ave Cyto |
Ch2 sd Cyto |
Cross ave Cyto |
Cross sd Cyto |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 1.112E+2 | 4.974E+1 | 1.237E+3 | 5.433E+2 | 1.034E+1 | 8.259E+0 | 2.354E+1 | 2.177E+1 | 2.107E+2 | 1.972E+2 | 1.407E+1 | 8.876E+0 |
| N_corr | 1.197E+2 | 5.491E+1 | 1.792E+3 | 9.087E+2 | 1.221E+1 | 1.009E+1 | 2.471E+1 | 2.733E+1 | 2.814E+2 | 3.912E+2 | 2.030E+1 | 1.658E+1 |
| c [nM] | 6.916E+2 | 3.094E+2 | 5.738E+3 | 2.520E+3 | 5.317E+1 | 4.246E+1 | 1.464E+2 | 1.354E+2 | 9.771E+2 | 9.147E+2 | 7.235E+1 | 4.563E+1 |
| c_corr [nM] | 7.446E+2 | 3.415E+2 | 8.313E+3 | 4.215E+3 | 6.276E+1 | 5.185E+1 | 1.537E+2 | 1.700E+2 | 1.305E+3 | 1.814E+3 | 1.044E+2 | 8.525E+1 |
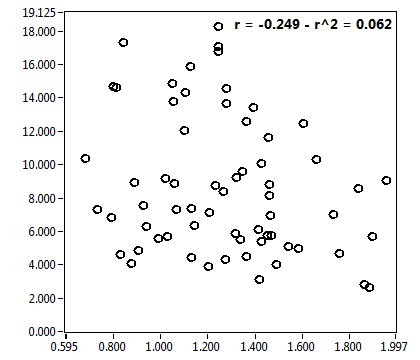
| ratioG | 8.252E-2 | 4.500E-2 | 4.157E-1 | 2.426E-1 | ||||||||
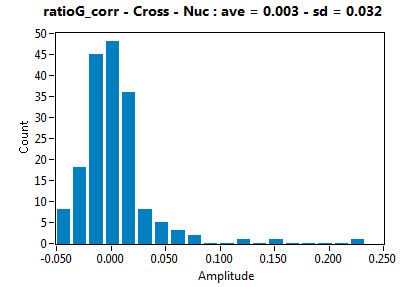
| ratioG_corr | 3.179E-3 | 3.200E-2 | 1.014E-1 | 7.935E-2 | ||||||||
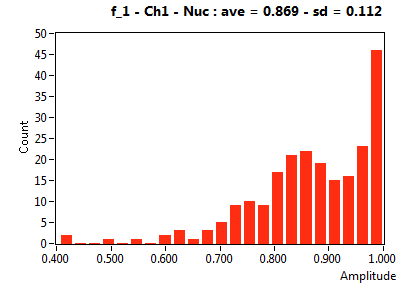
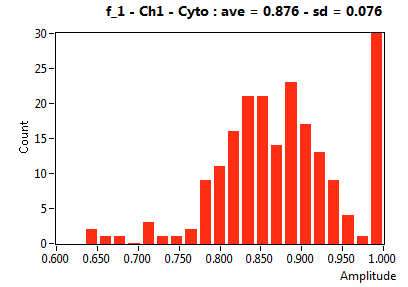
| f_1 | 8.691E-1 | 1.120E-1 | 3.644E-1 | 1.952E-1 | 9.668E-1 | 7.374E-2 | 8.760E-1 | 7.643E-2 | 6.748E-1 | 2.649E-1 | 9.173E-1 | 9.052E-2 |
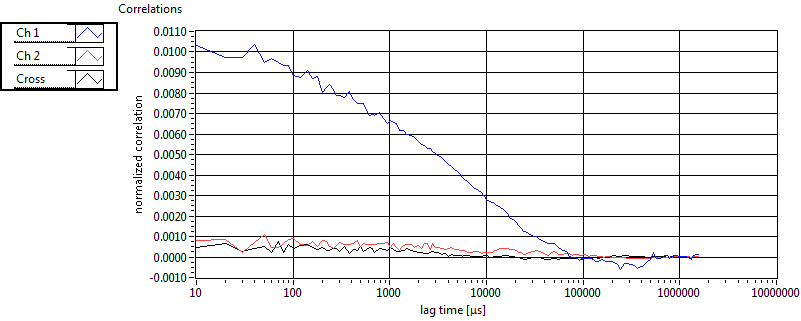
| tauD_1 [ms] | 2.608E+0 | 1.638E+0 | 1.860E+0 | 4.441E-16 | 2.224E+0 | 3.885E+0 | 1.349E+0 | 1.035E+0 | 1.860E+0 | 2.220E-16 | 1.479E+0 | 7.225E-1 |
| D_1 [mum^2/s] | 5.965E+0 | 4.105E+0 | 6.926E+0 | 8.882E-16 | 1.203E+1 | 1.609E+1 | 1.286E+1 | 1.054E+1 | 6.926E+0 | 0.000E+0 | 1.038E+1 | 5.854E+0 |
| alpha_1 | 9.458E-1 | 8.969E-2 | 9.203E-1 | 7.030E-4 | 8.605E-1 | 2.088E-1 | 9.170E-1 | 1.264E-1 | 9.202E-1 | 5.528E-4 | 7.789E-1 | 2.000E-1 |
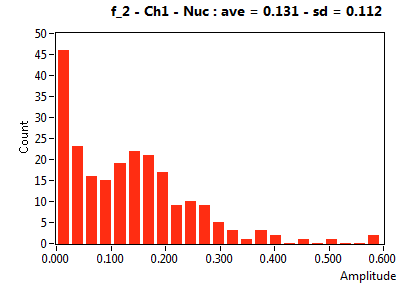
| f_2 | 1.309E-1 | 1.120E-1 | 6.356E-1 | 1.952E-1 | 3.319E-2 | 7.374E-2 | 1.240E-1 | 7.643E-2 | 3.252E-1 | 2.649E-1 | 8.265E-2 | 9.052E-2 |
| tauD_2 [ms] | 3.134E+1 | 1.860E+1 | 7.770E+1 | 8.423E+1 | 1.158E+2 | 2.582E+2 | 3.037E+1 | 3.414E+1 | 7.332E+1 | 8.032E+1 | 5.387E+1 | 3.872E+1 |
| D_2 [mum^2/s] | 4.122E-1 | 1.620E-1 | 5.126E-1 | 6.872E-1 | 1.842E-1 | 2.692E-1 | 5.277E-1 | 3.081E-1 | 5.488E-1 | 7.492E-1 | 2.967E-1 | 1.295E-1 |
| alpha_2 | 1.357E+0 | 2.857E-1 | 1.289E+0 | 2.873E-1 | 1.152E+0 | 2.118E-1 | 1.211E+0 | 2.097E-1 | 1.194E+0 | 3.138E-1 | 1.293E+0 | 3.084E-1 |
| tauD_m [ms] | 6.381E+0 | 5.311E+0 | 4.951E+1 | 6.133E+1 | 8.607E+0 | 2.645E+1 | 5.427E+0 | 1.182E+1 | 2.460E+1 | 4.122E+1 | 6.379E+0 | 9.625E+0 |
| D_m [mum^2/s] | 5.061E+0 | 3.260E+0 | 2.904E+0 | 1.165E+0 | 1.150E+1 | 1.484E+1 | 1.132E+1 | 9.581E+0 | 4.964E+0 | 1.474E+0 | 9.499E+0 | 5.265E+0 |
| R [1/s] | 1.865E+1 | 1.999E+1 | 1.782E+1 | 2.813E+1 | ||||||||
| CPM [kHz] | 1.547E+0 | 5.862E-1 | 4.128E-1 | 3.524E-1 | 1.143E+0 | 4.490E-1 | 2.621E-1 | 3.588E-1 | ||||
| CPM_corr [kHz] | 1.567E+0 | 5.859E-1 | 4.728E-1 | 3.977E-1 | 1.272E+0 | 4.515E-1 | 6.231E-1 | 4.502E-1 | ||||
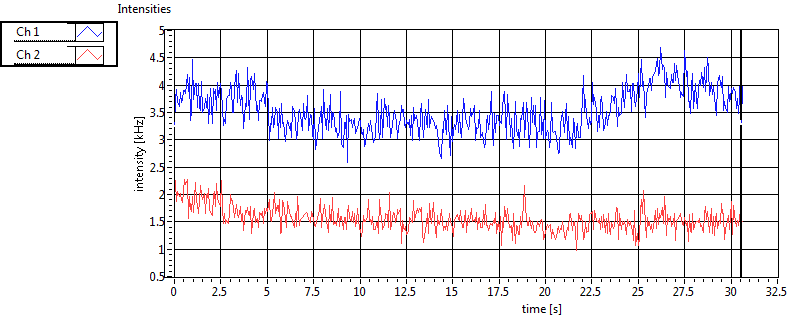
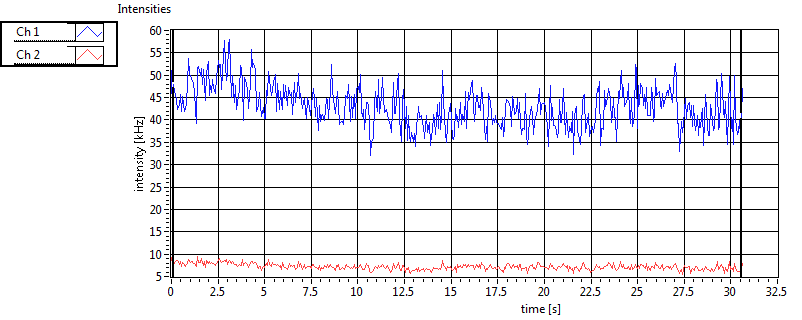
| Int [kHz] | 1.863E+2 | 1.018E+2 | 2.214E+2 | 6.112E+1 | 2.877E+1 | 2.532E+1 | 4.453E+1 | 5.579E+1 | ||||
| Int_corr [kHz] | 2.023E+2 | 1.111E+2 | 3.623E+2 | 1.088E+2 | 3.243E+1 | 3.096E+1 | 7.861E+1 | 1.136E+2 | ||||
| # samples | 225 | 225 | 187 | 187 | 176 | 176 | 200 | 200 | 36 | 36 | 29 | 29 |
| # cells | 80 |
![c_corr [nM] - Ch1 - Nuc : ave = 744.632 - sd = 341.518](1909_c_corr_1n.png)
![Int_corr [kHz] - Ch1 - Nuc : ave = 202.267 - sd = 111.118](1909_Int_corr_1n.png)
![D_1 [mum^2/s] - Ch1 - Nuc : ave = 5.965 - sd = 4.105](1909_D_1_1n.png)
![D_2 [mum^2/s] - Ch1 - Nuc : ave = 0.412 - sd = 0.162](1909_D_2_1n.png)
![CPM_corr [kHz] - Ch1 - Nuc : ave = 1.567 - sd = 0.586](1909_CPM_corr_1n.png)
![D_m [mum^2/s] - Ch1 - Nuc : ave = 5.061 - sd = 3.260](1909_D_m_1n.png)

![c_corr [nM] - Ch1 - Nuc : ave = 744.632 - sd = 341.518](1909_c_corr_1n.png)
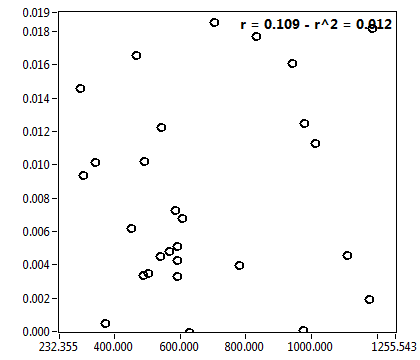
![c_corr [nM] - Ch1 - Cyto : ave = 153.707 - sd = 169.988](1909_c_corr_1c.png)
![Int_corr [kHz] - Ch1 - Cyto : ave = 32.425 - sd = 30.959](1909_Int_corr_1c.png)
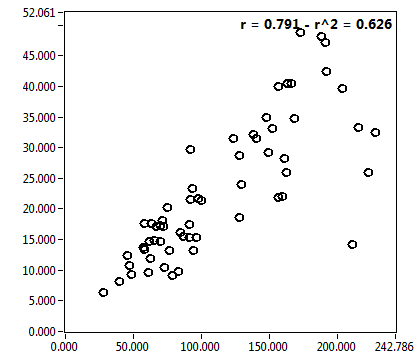
![D_1 [mum^2/s] - Ch1 - Cyto : ave = 12.857 - sd = 10.539](1909_D_1_1c.png)
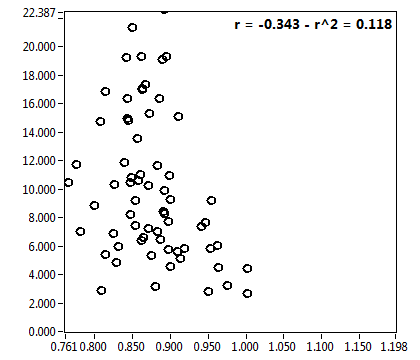
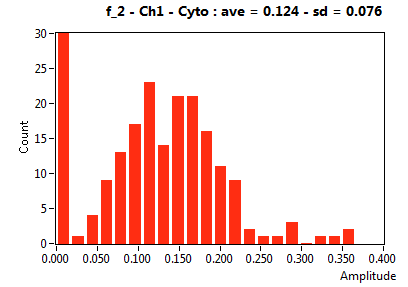
![D_2 [mum^2/s] - Ch1 - Cyto : ave = 0.528 - sd = 0.308](1909_D_2_1c.png)
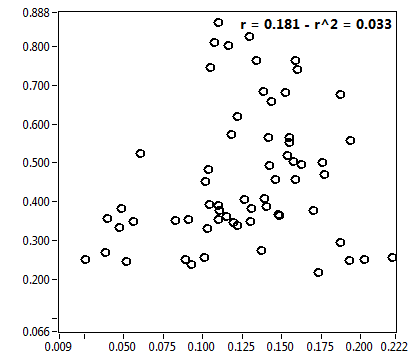
![CPM_corr [kHz] - Ch1 - Cyto : ave = 1.272 - sd = 0.451](1909_CPM_corr_1c.png)
![D_m [mum^2/s] - Ch1 - Cyto : ave = 11.324 - sd = 9.581](1909_D_m_1c.png)