 |  |
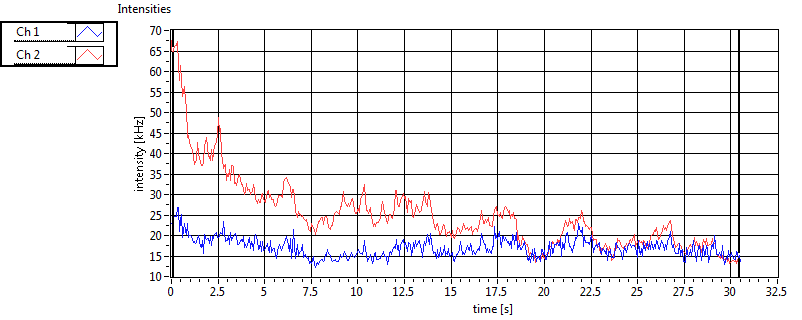
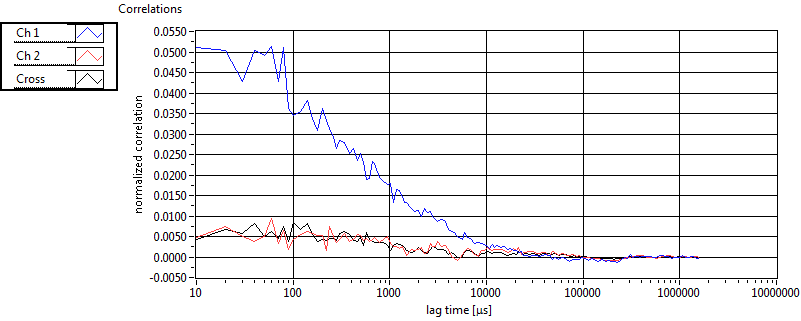
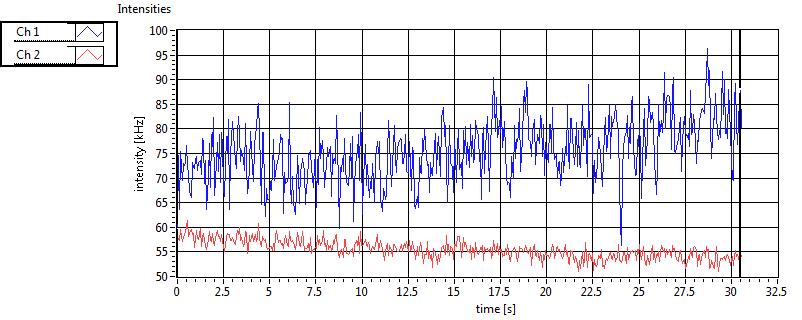
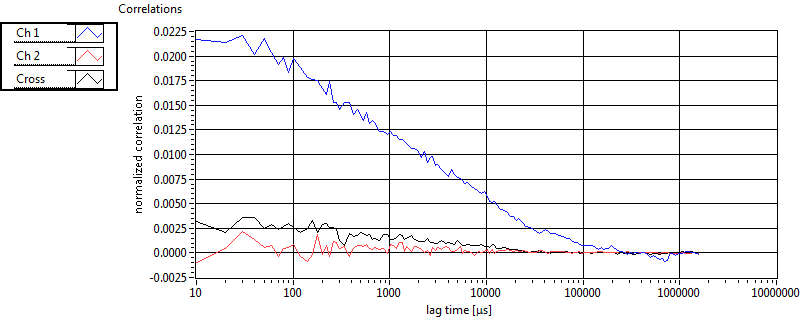
| 1 | 20110915\U01--V01--1947 |
|---|---|
| 2 | 20110916\U01--V01--1947 |
| 3 | 20110920\U01--V00--1947 |
| 4 | 20111208\U01--V00--1947 |
| 5 | 20120116\U01--V00--1947 |
| 6 | 20120127\U00--V00--1947 |
| Ch1 ave Nuc |
Ch1 sd Nuc |
Ch2 ave Nuc |
Ch2 sd Nuc |
Cross ave Nuc |
Cross sd Nuc |
Ch1 ave Cyto |
Ch1 sd Cyto |
Ch2 ave Cyto |
Ch2 sd Cyto |
Cross ave Cyto |
Cross sd Cyto |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 8.713E+1 | 6.664E+1 | 1.101E+3 | 6.770E+2 | 9.834E+0 | 1.860E+1 | 2.735E+1 | 3.284E+1 | 5.509E+2 | 3.056E+2 | 2.220E+1 | 3.380E+1 |
| N_corr | 9.220E+1 | 6.845E+1 | 1.591E+3 | 1.138E+3 | 1.217E+1 | 2.695E+1 | 2.795E+1 | 3.681E+1 | 7.124E+2 | 5.332E+2 | 2.184E+1 | 3.254E+1 |
| c [nM] | 5.419E+2 | 4.144E+2 | 5.108E+3 | 3.140E+3 | 5.055E+1 | 9.563E+1 | 1.701E+2 | 2.043E+2 | 2.555E+3 | 1.417E+3 | 1.141E+2 | 1.738E+2 |
| c_corr [nM] | 5.734E+2 | 4.257E+2 | 7.379E+3 | 5.278E+3 | 6.256E+1 | 1.385E+2 | 1.738E+2 | 2.289E+2 | 3.304E+3 | 2.473E+3 | 1.123E+2 | 1.673E+2 |
| ratioG | 8.475E-2 | 7.744E-2 | 2.623E-1 | 2.200E-1 | ||||||||
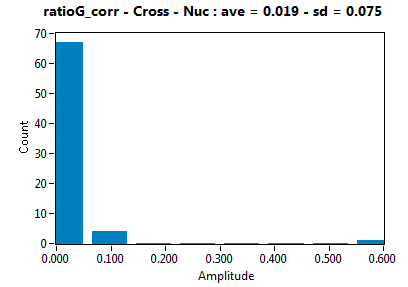
| ratioG_corr | 1.881E-2 | 7.494E-2 | 9.416E-2 | 1.300E-1 | ||||||||
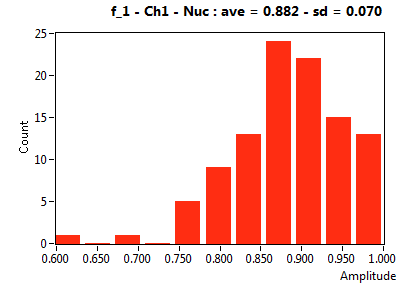
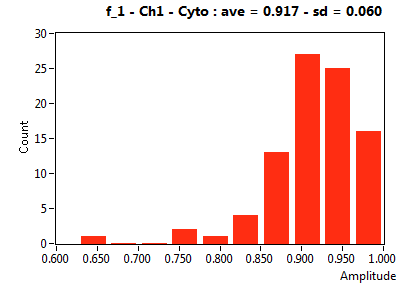
| f_1 | 8.820E-1 | 6.976E-2 | 3.614E-1 | 2.250E-1 | 9.737E-1 | 6.477E-2 | 9.172E-1 | 6.020E-2 | 7.036E-1 | 2.804E-1 | 9.447E-1 | 4.040E-2 |
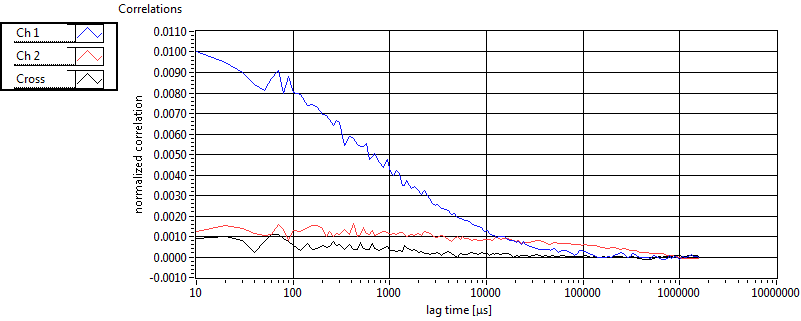
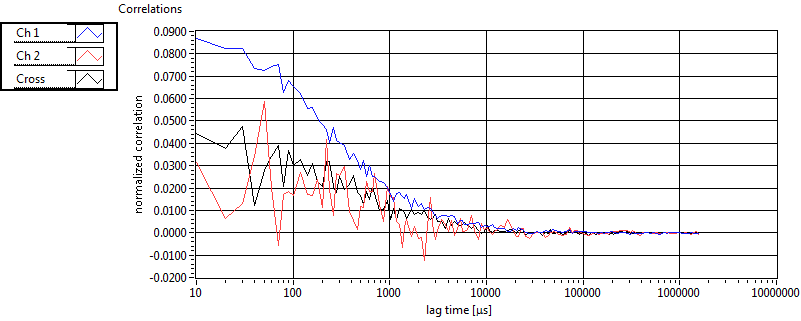
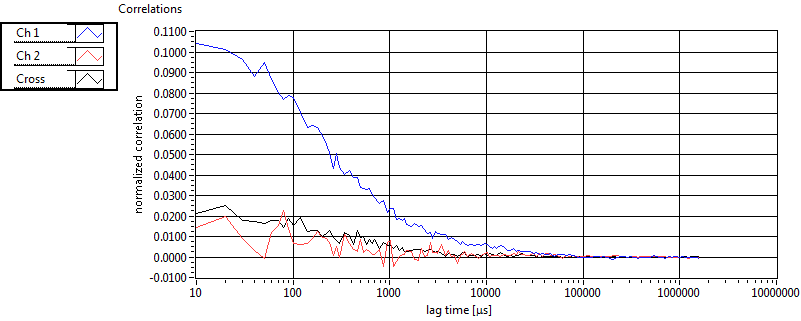
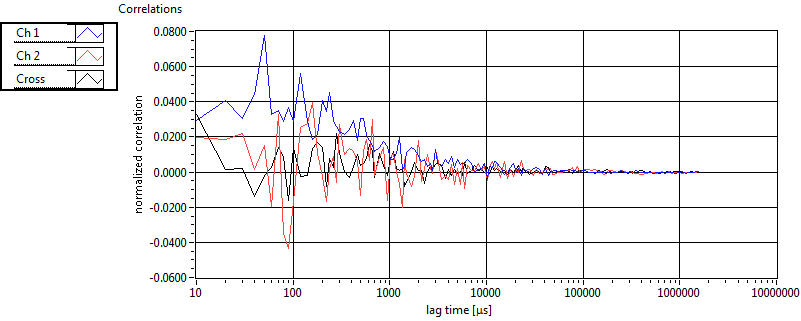
| tauD_1 [ms] | 1.369E+0 | 5.710E-1 | 1.839E+0 | 2.867E-2 | 1.060E+0 | 7.620E-1 | 6.295E-1 | 4.604E-1 | 2.453E+0 | 1.551E+0 | 1.360E+0 | 9.286E-1 |
| D_1 [mum^2/s] | 9.102E+0 | 3.778E+0 | 7.007E+0 | 1.103E-1 | 1.914E+1 | 1.768E+1 | 2.379E+1 | 1.158E+1 | 6.268E+0 | 1.664E+0 | 1.890E+1 | 1.735E+1 |
| alpha_1 | 9.489E-1 | 7.488E-2 | 9.200E-1 | 0.000E+0 | 8.933E-1 | 2.764E-1 | 1.029E+0 | 1.226E-1 | 9.031E-1 | 4.232E-2 | 1.105E+0 | 1.824E-1 |
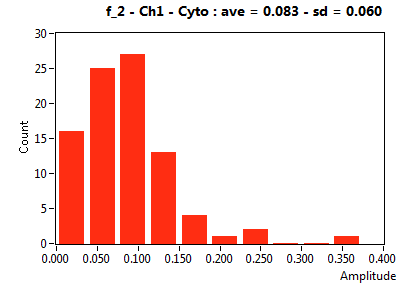
| f_2 | 1.180E-1 | 6.976E-2 | 6.386E-1 | 2.250E-1 | 2.628E-2 | 6.477E-2 | 8.282E-2 | 6.020E-2 | 2.964E-1 | 2.804E-1 | 5.531E-2 | 4.040E-2 |
| tauD_2 [ms] | 2.144E+1 | 7.243E+0 | 8.471E+1 | 8.203E+1 | 7.812E+1 | 4.649E+1 | 1.871E+1 | 2.752E+1 | 3.279E+1 | 3.364E+1 | 4.751E+1 | 2.556E+1 |
| D_2 [mum^2/s] | 5.443E-1 | 1.638E-1 | 4.869E-1 | 7.750E-1 | 2.017E-1 | 9.813E-2 | 9.643E-1 | 5.330E-1 | 1.794E+0 | 2.212E+0 | 4.217E-1 | 3.854E-1 |
| alpha_2 | 1.299E+0 | 2.214E-1 | 1.302E+0 | 3.187E-1 | 1.093E+0 | 9.648E-2 | 1.103E+0 | 1.600E-1 | 1.136E+0 | 2.734E-1 | 1.049E+0 | 2.568E-1 |
| tauD_m [ms] | 3.766E+0 | 2.368E+0 | 5.281E+1 | 5.406E+1 | 3.682E+0 | 7.609E+0 | 2.277E+0 | 3.721E+0 | 1.112E+1 | 1.125E+1 | 3.654E+0 | 2.877E+0 |
| D_m [mum^2/s] | 8.076E+0 | 3.249E+0 | 2.875E+0 | 1.479E+0 | 1.878E+1 | 1.763E+1 | 2.195E+1 | 1.092E+1 | 4.855E+0 | 1.656E+0 | 1.755E+1 | 1.567E+1 |
| R [1/s] | 3.647E+1 | 2.580E+1 | 3.412E+1 | 4.070E+1 | ||||||||
| CPM [kHz] | 1.338E+0 | 3.618E-1 | 3.418E-1 | 2.777E-1 | 9.178E-1 | 4.705E-1 | 1.994E-1 | 7.477E-2 | ||||
| CPM_corr [kHz] | 1.370E+0 | 3.629E-1 | 3.862E-1 | 3.102E-1 | 1.088E+0 | 5.010E-1 | 3.249E-1 | 2.479E-1 | ||||
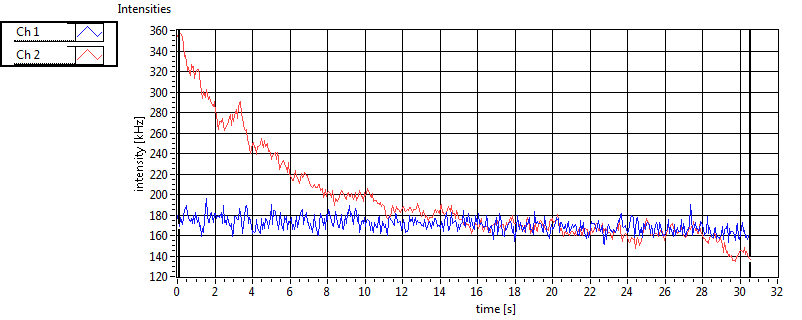
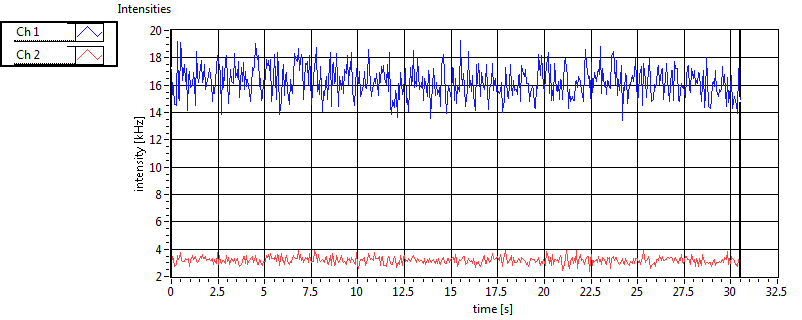
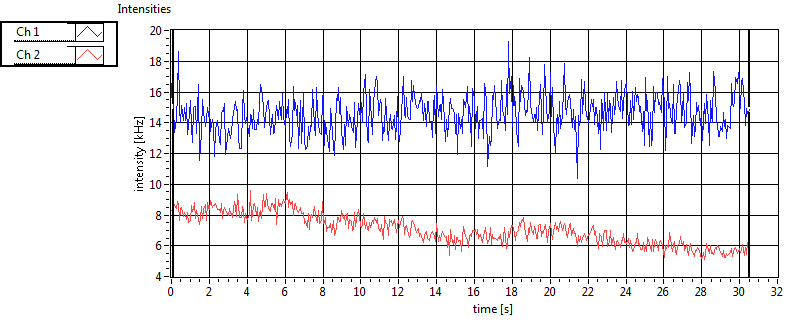
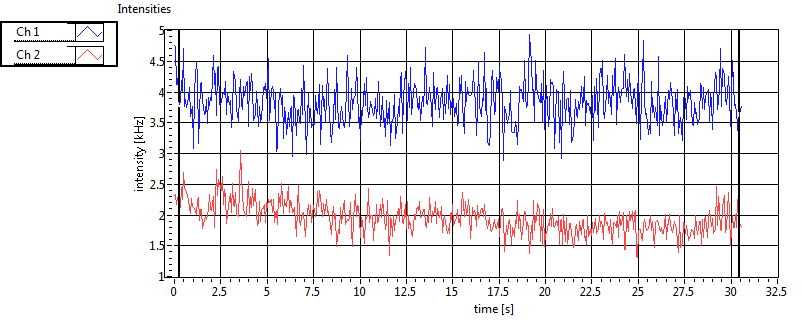
| Int [kHz] | 1.213E+2 | 7.936E+1 | 1.780E+2 | 7.617E+1 | 2.339E+1 | 3.154E+1 | 8.807E+1 | 4.478E+1 | ||||
| Int_corr [kHz] | 1.308E+2 | 8.517E+1 | 2.832E+2 | 1.494E+2 | 2.526E+1 | 3.221E+1 | 1.298E+2 | 8.830E+1 | ||||
| # samples | 103 | 103 | 85 | 85 | 72 | 72 | 89 | 89 | 13 | 13 | 9 | 9 |
| # cells | 41 |
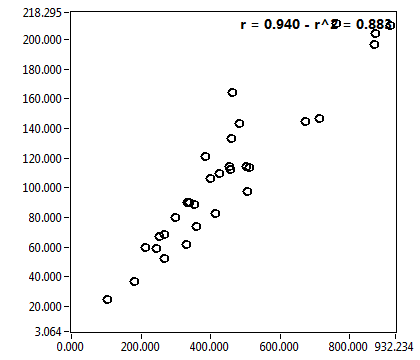
![c_corr [nM] - Ch1 - Nuc : ave = 573.421 - sd = 425.734](1947_c_corr_1n.png)
![Int_corr [kHz] - Ch1 - Nuc : ave = 130.816 - sd = 85.168](1947_Int_corr_1n.png)
![D_1 [mum^2/s] - Ch1 - Nuc : ave = 9.102 - sd = 3.778](1947_D_1_1n.png)
![D_2 [mum^2/s] - Ch1 - Nuc : ave = 0.544 - sd = 0.164](1947_D_2_1n.png)
![CPM_corr [kHz] - Ch1 - Nuc : ave = 1.370 - sd = 0.363](1947_CPM_corr_1n.png)
![D_m [mum^2/s] - Ch1 - Nuc : ave = 8.076 - sd = 3.249](1947_D_m_1n.png)
![c_corr [nM] - Ch1 - Nuc : ave = 573.421 - sd = 425.734](1947_c_corr_1n.png)

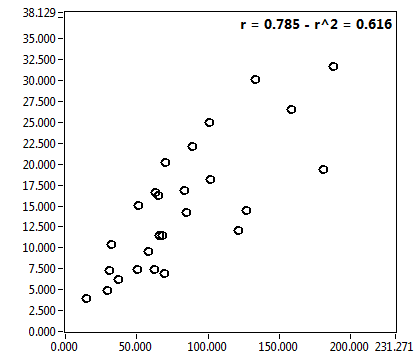
![c_corr [nM] - Ch1 - Cyto : ave = 173.818 - sd = 228.907](1947_c_corr_1c.png)
![Int_corr [kHz] - Ch1 - Cyto : ave = 25.260 - sd = 32.209](1947_Int_corr_1c.png)
![D_1 [mum^2/s] - Ch1 - Cyto : ave = 23.792 - sd = 11.580](1947_D_1_1c.png)
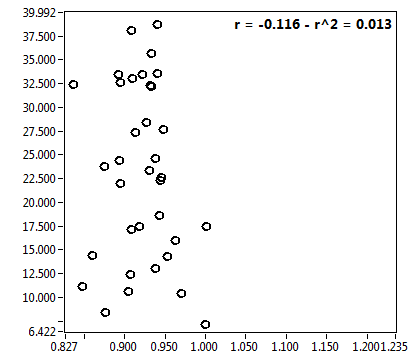
![D_2 [mum^2/s] - Ch1 - Cyto : ave = 0.964 - sd = 0.533](1947_D_2_1c.png)
![CPM_corr [kHz] - Ch1 - Cyto : ave = 1.088 - sd = 0.501](1947_CPM_corr_1c.png)
![D_m [mum^2/s] - Ch1 - Cyto : ave = 21.945 - sd = 10.915](1947_D_m_1c.png)
