 |  |
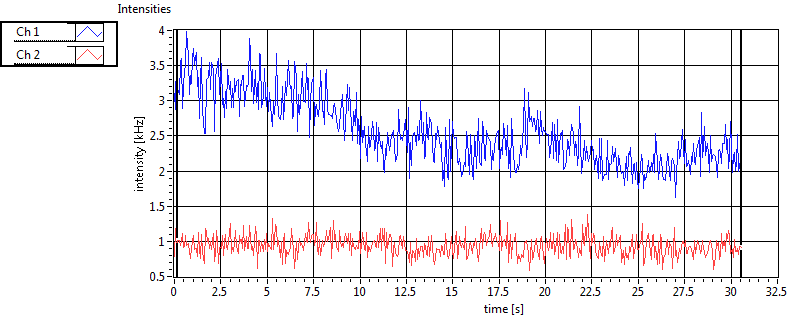
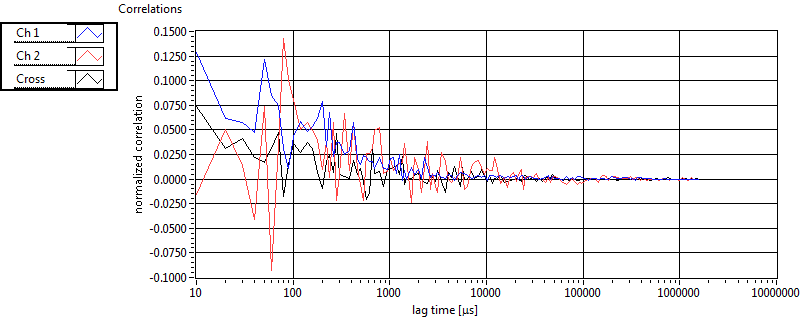
| 1 | 20130109\U04--V00--2018hi |
|---|---|
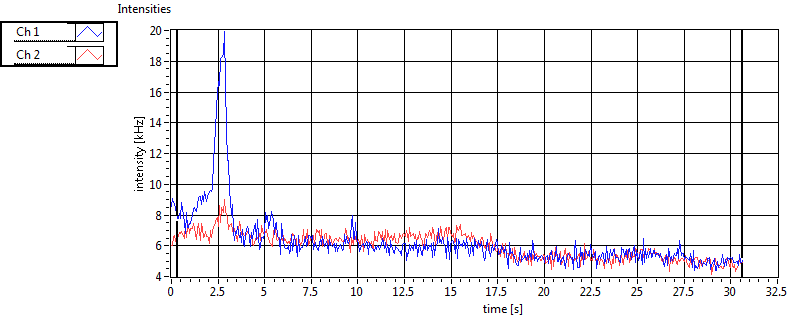
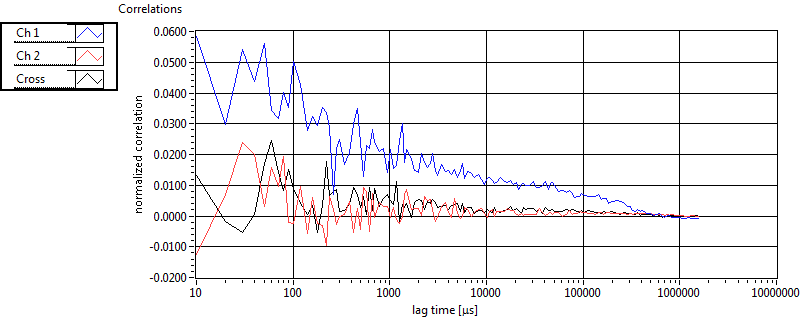
| 2 | 20130111\U01--V00--2018hi |
| Ch1 ave Nuc |
Ch1 sd Nuc |
Ch2 ave Nuc |
Ch2 sd Nuc |
Cross ave Nuc |
Cross sd Nuc |
Ch1 ave Cyto |
Ch1 sd Cyto |
Ch2 ave Cyto |
Ch2 sd Cyto |
Cross ave Cyto |
Cross sd Cyto |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 7.351E+2 | 4.367E+2 | 1.077E+3 | 5.321E+2 | 2.709E+2 | 1.915E+2 | 1.096E+2 | 2.926E+2 | 8.767E+2 | 3.168E+2 | 2.803E+2 | 0.000E+0 |
| N_corr | 7.489E+2 | 4.433E+2 | 1.020E+3 | 5.026E+2 | 2.837E+2 | 1.983E+2 | 1.874E+2 | 5.420E+2 | 9.638E+2 | 3.821E+2 | 2.591E+2 | 0.000E+0 |
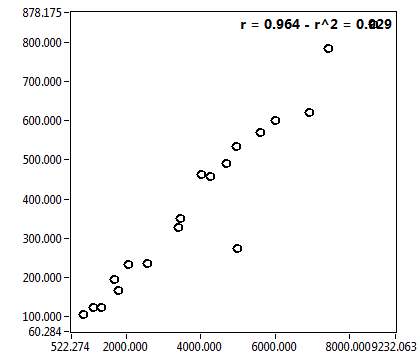
| c [nM] | 4.572E+3 | 2.716E+3 | 4.996E+3 | 2.468E+3 | 1.393E+3 | 9.845E+2 | 6.816E+2 | 1.820E+3 | 4.066E+3 | 1.469E+3 | 1.441E+3 | 0.000E+0 |
| c_corr [nM] | 4.658E+3 | 2.757E+3 | 4.731E+3 | 2.331E+3 | 1.459E+3 | 1.019E+3 | 1.165E+3 | 3.371E+3 | 4.470E+3 | 1.772E+3 | 1.332E+3 | 0.000E+0 |
| ratioG | 2.755E-1 | 1.050E-1 | 3.716E-1 | 0.000E+0 | ||||||||
| ratioG_corr | 5.029E-2 | 8.387E-2 | 8.279E-2 | 0.000E+0 | ||||||||
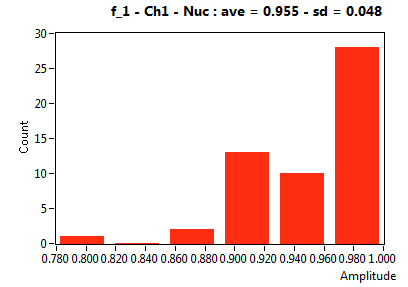
| f_1 | 9.551E-1 | 4.774E-2 | 4.969E-1 | 2.974E-1 | 9.924E-1 | 2.627E-2 | 9.463E-1 | 7.014E-2 | 6.386E-1 | 1.638E-1 | 1.000E+0 | 0.000E+0 |
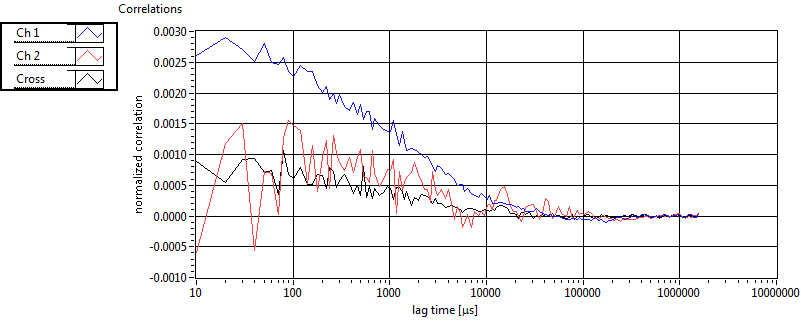
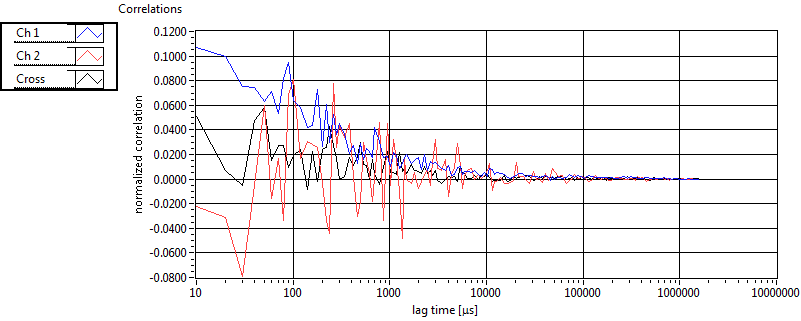
| tauD_1 [ms] | 1.210E+0 | 9.831E-1 | 1.860E+0 | 0.000E+0 | 5.180E-1 | 4.631E-1 | 5.134E-1 | 4.799E-1 | 1.860E+0 | 0.000E+0 | 3.768E-1 | 0.000E+0 |
| D_1 [mum^2/s] | 1.244E+1 | 5.532E+0 | 6.926E+0 | 0.000E+0 | 4.137E+1 | 3.027E+1 | 3.526E+1 | 2.795E+1 | 6.926E+0 | 0.000E+0 | 3.182E+1 | 0.000E+0 |
| alpha_1 | 1.046E+0 | 9.629E-2 | 9.200E-1 | 1.110E-16 | 9.974E-1 | 1.930E-1 | 1.087E+0 | 1.395E-1 | 9.200E-1 | 0.000E+0 | 7.503E-1 | 0.000E+0 |
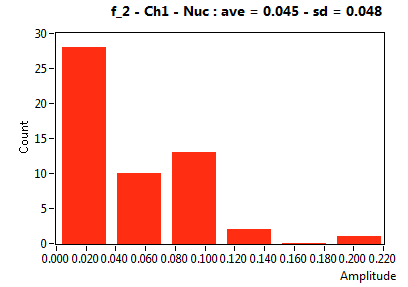
| f_2 | 4.490E-2 | 4.774E-2 | 5.031E-1 | 2.974E-1 | 7.582E-3 | 2.627E-2 | 5.369E-2 | 7.014E-2 | 3.614E-1 | 1.638E-1 | 0.000E+0 | 0.000E+0 |
| tauD_2 [ms] | 1.691E+1 | 9.200E+0 | 4.348E+1 | 3.797E+1 | 4.068E+1 | 1.616E+1 | 1.747E+1 | 1.705E+1 | 1.782E+1 | 2.094E+0 | 1.987E+1 | 0.000E+0 |
| D_2 [mum^2/s] | 7.623E-1 | 2.934E-1 | 7.151E-1 | 7.345E-1 | 3.382E-1 | 1.182E-1 | 9.647E-1 | 6.417E-1 | 7.329E-1 | 8.648E-2 | 6.033E-1 | 0.000E+0 |
| alpha_2 | 1.128E+0 | 1.271E-1 | 1.169E+0 | 3.327E-1 | 1.087E+0 | 5.046E-2 | 1.100E+0 | 5.091E-2 | 1.119E+0 | 1.955E-1 | 1.077E+0 | 0.000E+0 |
| tauD_m [ms] | 1.927E+0 | 1.221E+0 | 1.735E+1 | 1.529E+1 | 7.629E-1 | 8.921E-1 | 2.181E+0 | 5.564E+0 | 7.509E+0 | 2.361E+0 | 3.768E-1 | 0.000E+0 |
| D_m [mum^2/s] | 1.186E+1 | 5.263E+0 | 3.918E+0 | 1.695E+0 | 4.104E+1 | 3.006E+1 | 3.335E+1 | 2.693E+1 | 4.692E+0 | 1.002E+0 | 3.182E+1 | 0.000E+0 |
| R [1/s] | 4.588E+1 | 4.629E+1 | 3.900E+1 | 0.000E+0 | ||||||||
| CPM [kHz] | 5.873E-1 | 9.340E-2 | 2.398E-1 | 1.245E-1 | 2.796E-1 | 1.337E-1 | 2.253E-1 | 8.418E-2 | ||||
| CPM_corr [kHz] | 5.915E-1 | 9.309E-2 | 3.418E-1 | 1.597E-1 | 4.738E-1 | 2.559E-1 | 2.935E-1 | 6.519E-2 | ||||
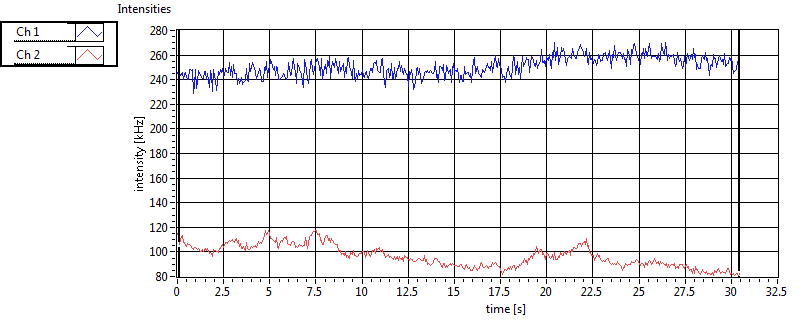
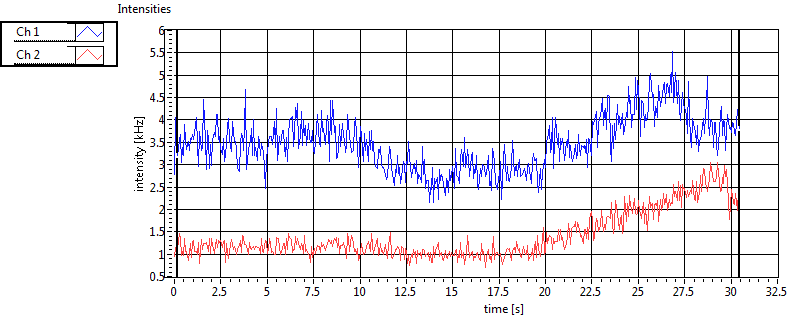
| Int [kHz] | 4.574E+2 | 2.673E+2 | 1.923E+2 | 4.913E+1 | 4.468E+1 | 1.219E+2 | 1.519E+2 | 1.818E+1 | ||||
| Int_corr [kHz] | 4.682E+2 | 2.719E+2 | 2.717E+2 | 7.492E+1 | 7.173E+1 | 1.869E+2 | 2.309E+2 | 4.667E+1 | ||||
| # samples | 54 | 54 | 41 | 41 | 32 | 32 | 34 | 34 | 4 | 4 | 1 | 1 |
| # cells | 33 |
![c_corr [nM] - Ch1 - Nuc : ave = 4657.654 - sd = 2756.920](2018hi_c_corr_1n.png)
![Int_corr [kHz] - Ch1 - Nuc : ave = 468.164 - sd = 271.920](2018hi_Int_corr_1n.png)
![D_1 [mum^2/s] - Ch1 - Nuc : ave = 12.443 - sd = 5.532](2018hi_D_1_1n.png)
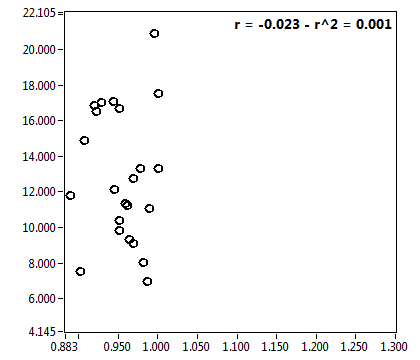
![D_2 [mum^2/s] - Ch1 - Nuc : ave = 0.762 - sd = 0.293](2018hi_D_2_1n.png)
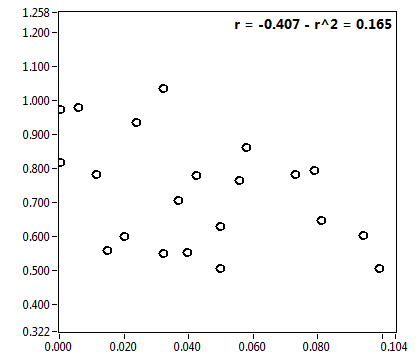
![CPM_corr [kHz] - Ch1 - Nuc : ave = 0.592 - sd = 0.093](2018hi_CPM_corr_1n.png)
![D_m [mum^2/s] - Ch1 - Nuc : ave = 11.862 - sd = 5.263](2018hi_D_m_1n.png)
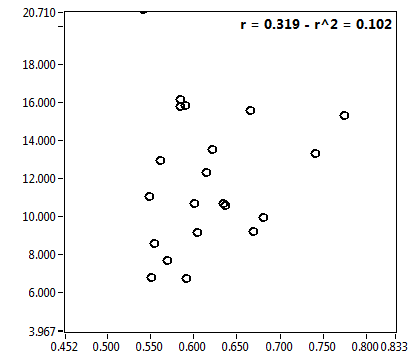
![c_corr [nM] - Ch1 - Nuc : ave = 4657.654 - sd = 2756.920](2018hi_c_corr_1n.png)
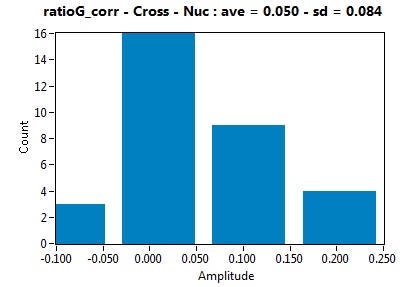

![c_corr [nM] - Ch1 - Cyto : ave = 1165.326 - sd = 3370.996](2018hi_c_corr_1c.png)
![Int_corr [kHz] - Ch1 - Cyto : ave = 71.726 - sd = 186.935](2018hi_Int_corr_1c.png)
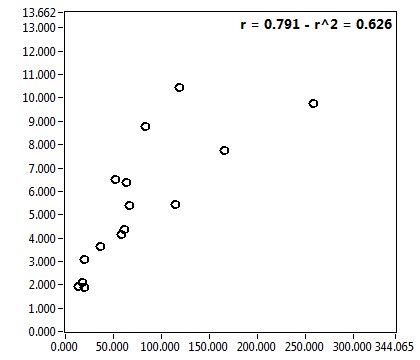
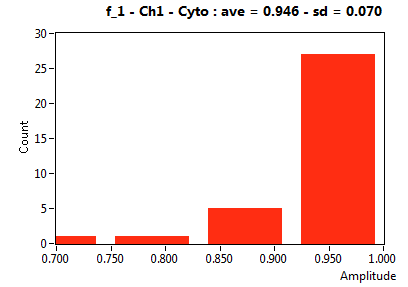
![D_1 [mum^2/s] - Ch1 - Cyto : ave = 35.258 - sd = 27.953](2018hi_D_1_1c.png)
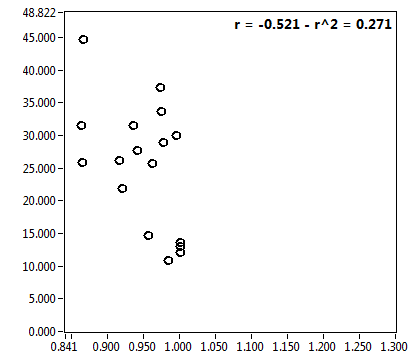
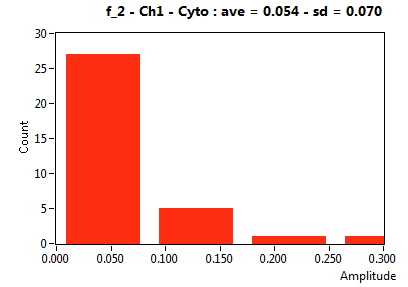
![D_2 [mum^2/s] - Ch1 - Cyto : ave = 0.965 - sd = 0.642](2018hi_D_2_1c.png)
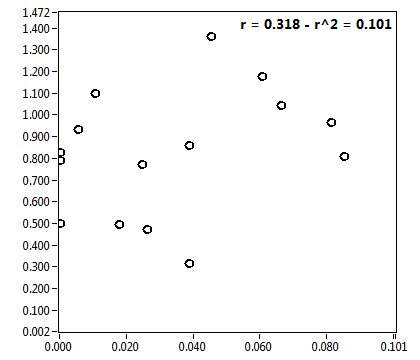
![CPM_corr [kHz] - Ch1 - Cyto : ave = 0.474 - sd = 0.256](2018hi_CPM_corr_1c.png)
![D_m [mum^2/s] - Ch1 - Cyto : ave = 33.352 - sd = 26.930](2018hi_D_m_1c.png)
