 |  |
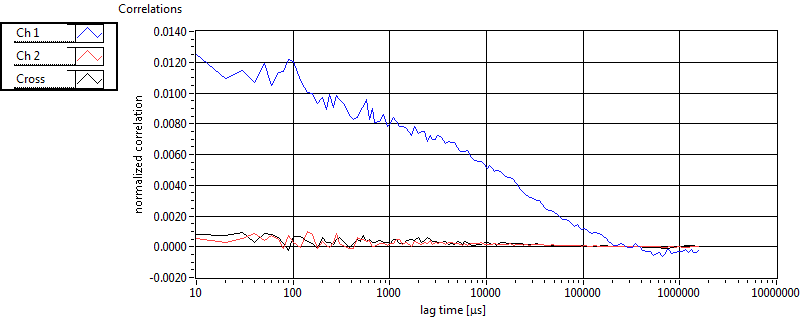
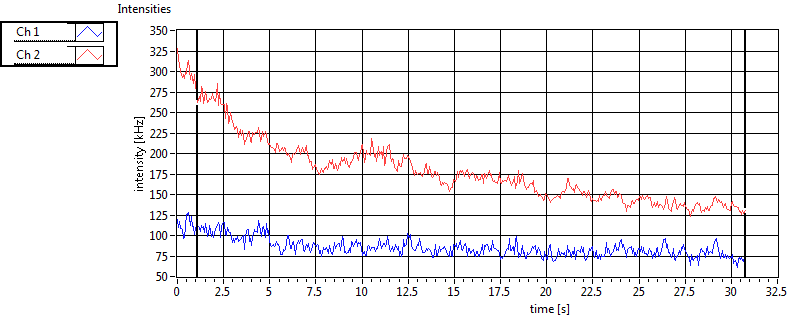
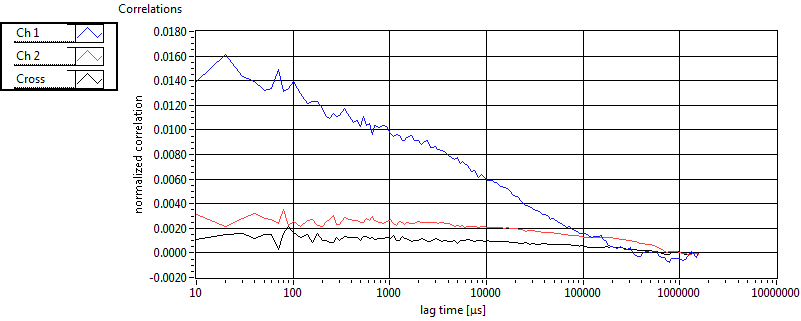
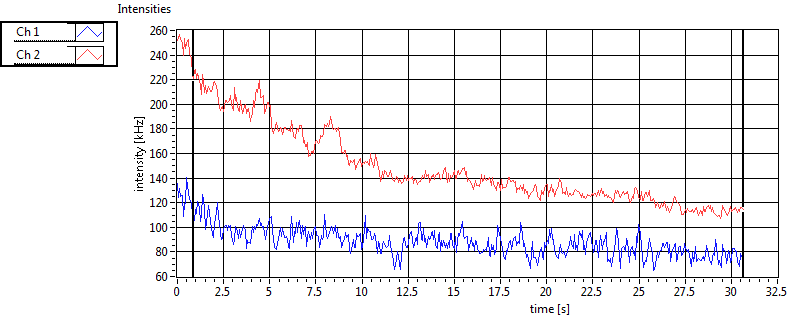
| 1 | 20111117\U03--V00--2093 |
|---|---|
| 2 | 20111206\U01--V01--2093 |
| 3 | 20111207\U02--V01--2093 |
| 4 | 20111216\U02--V00--2093 |
| Ch1 ave Nuc |
Ch1 sd Nuc |
Ch2 ave Nuc |
Ch2 sd Nuc |
Cross ave Nuc |
Cross sd Nuc |
Ch1 ave Cyto |
Ch1 sd Cyto |
Ch2 ave Cyto |
Ch2 sd Cyto |
Cross ave Cyto |
Cross sd Cyto |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 1.328E+2 | 8.212E+1 | 1.293E+3 | 6.137E+2 | 1.308E+1 | 1.606E+1 | 4.232E+1 | 6.493E+1 | 4.156E+2 | 2.589E+2 | 3.759E+1 | 4.672E+1 |
| N_corr | 1.805E+2 | 1.172E+2 | 1.992E+3 | 1.077E+3 | 2.525E+1 | 3.347E+1 | 4.636E+1 | 7.656E+1 | 4.822E+2 | 3.659E+2 | 7.375E+1 | 7.998E+1 |
| c [nM] | 8.261E+2 | 5.107E+2 | 5.999E+3 | 2.847E+3 | 6.722E+1 | 8.258E+1 | 2.632E+2 | 4.038E+2 | 1.928E+3 | 1.201E+3 | 1.933E+2 | 2.402E+2 |
| c_corr [nM] | 1.122E+3 | 7.287E+2 | 9.239E+3 | 4.997E+3 | 1.298E+2 | 1.721E+2 | 2.884E+2 | 4.761E+2 | 2.237E+3 | 1.697E+3 | 3.792E+2 | 4.112E+2 |
| ratioG | 8.732E-2 | 6.218E-2 | 3.028E-1 | 1.819E-1 | ||||||||
| ratioG_corr | 5.136E-2 | 5.927E-2 | 2.367E-1 | 2.249E-1 | ||||||||
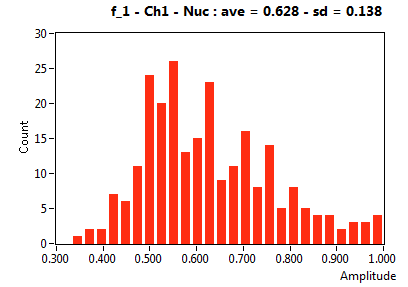
| f_1 | 6.279E-1 | 1.383E-1 | 4.115E-1 | 2.302E-1 | 8.884E-1 | 1.572E-1 | 9.348E-1 | 8.541E-2 | 7.115E-1 | 3.454E-1 | 9.523E-1 | 9.359E-2 |
| tauD_1 [ms] | 2.827E+0 | 2.187E+0 | 1.860E+0 | 4.441E-16 | 5.864E+0 | 8.176E+0 | 5.928E-1 | 7.085E-1 | 1.860E+0 | 0.000E+0 | 2.496E+0 | 1.316E+0 |
| D_1 [mum^2/s] | 7.800E+0 | 1.325E+1 | 6.926E+0 | 1.776E-15 | 9.186E+0 | 2.029E+1 | 3.851E+1 | 2.881E+1 | 6.926E+0 | 8.882E-16 | 8.381E+0 | 8.305E+0 |
| alpha_1 | 9.366E-1 | 1.086E-1 | 9.200E-1 | 4.441E-16 | 9.008E-1 | 2.514E-1 | 9.412E-1 | 1.526E-1 | 9.200E-1 | 0.000E+0 | 1.042E+0 | 2.129E-1 |
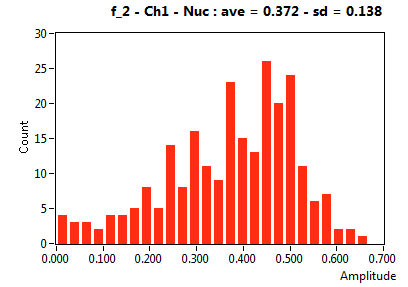
| f_2 | 3.721E-1 | 1.383E-1 | 5.885E-1 | 2.302E-1 | 1.116E-1 | 1.572E-1 | 6.522E-2 | 8.541E-2 | 2.885E-1 | 3.454E-1 | 4.771E-2 | 9.359E-2 |
| tauD_2 [ms] | 4.951E+1 | 3.827E+1 | 7.847E+1 | 9.140E+1 | 1.492E+2 | 1.239E+2 | 1.872E+1 | 2.451E+1 | 8.406E+1 | 1.568E+2 | 8.700E+1 | 1.044E+2 |
| D_2 [mum^2/s] | 4.152E-1 | 1.172E+0 | 5.823E-1 | 7.484E-1 | 1.166E-1 | 7.123E-2 | 1.005E+0 | 5.932E-1 | 1.068E+0 | 1.440E+0 | 2.956E-1 | 3.491E-1 |
| alpha_2 | 1.712E+0 | 3.060E-1 | 1.271E+0 | 2.997E-1 | 1.294E+0 | 3.473E-1 | 1.167E+0 | 2.000E-1 | 1.190E+0 | 3.015E-1 | 1.146E+0 | 2.358E-1 |
| tauD_m [ms] | 1.916E+1 | 1.166E+1 | 4.317E+1 | 5.427E+1 | 2.552E+1 | 4.148E+1 | 2.349E+0 | 5.017E+0 | 1.870E+1 | 4.638E+1 | 1.474E+1 | 3.188E+1 |
| D_m [mum^2/s] | 5.565E+0 | 1.257E+1 | 3.247E+0 | 1.397E+0 | 7.914E+0 | 1.716E+1 | 3.680E+1 | 2.849E+1 | 5.632E+0 | 1.445E+0 | 7.947E+0 | 8.080E+0 |
| R [1/s] | 1.053E+1 | 2.440E+1 | 2.410E+1 | 5.532E+1 | ||||||||
| CPM [kHz] | 9.004E-1 | 3.461E-1 | 4.489E-1 | 4.931E-1 | 6.395E-1 | 9.543E-1 | 2.900E-1 | 1.777E-1 | ||||
| CPM_corr [kHz] | 9.176E-1 | 3.486E-1 | 4.939E-1 | 5.448E-1 | 8.908E-1 | 1.015E+0 | 1.324E+0 | 3.853E+0 | ||||
| Int [kHz] | 1.165E+2 | 6.942E+1 | 2.399E+2 | 6.945E+1 | 2.029E+1 | 3.464E+1 | 9.393E+1 | 5.935E+1 | ||||
| Int_corr [kHz] | 1.571E+2 | 9.071E+1 | 3.964E+2 | 1.306E+2 | 2.843E+1 | 8.424E+1 | 1.311E+2 | 8.914E+1 | ||||
| # samples | 246 | 246 | 192 | 192 | 161 | 161 | 169 | 169 | 19 | 19 | 15 | 15 |
| # cells | 103 |
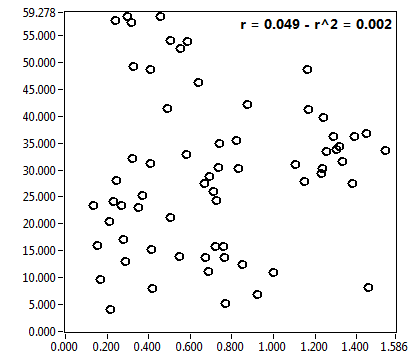
![c_corr [nM] - Ch1 - Nuc : ave = 1122.382 - sd = 728.655](2093_c_corr_1n.png)
![Int_corr [kHz] - Ch1 - Nuc : ave = 157.138 - sd = 90.715](2093_Int_corr_1n.png)
![D_1 [mum^2/s] - Ch1 - Nuc : ave = 7.800 - sd = 13.248](2093_D_1_1n.png)
![D_2 [mum^2/s] - Ch1 - Nuc : ave = 0.415 - sd = 1.172](2093_D_2_1n.png)
![CPM_corr [kHz] - Ch1 - Nuc : ave = 0.918 - sd = 0.349](2093_CPM_corr_1n.png)
![D_m [mum^2/s] - Ch1 - Nuc : ave = 5.565 - sd = 12.568](2093_D_m_1n.png)
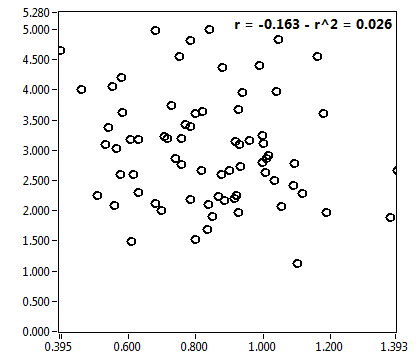
![c_corr [nM] - Ch1 - Nuc : ave = 1122.382 - sd = 728.655](2093_c_corr_1n.png)
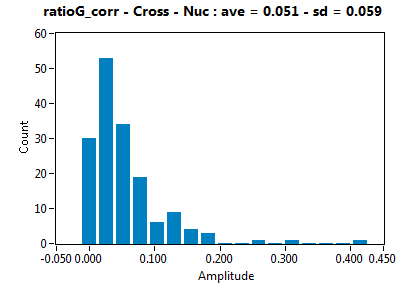
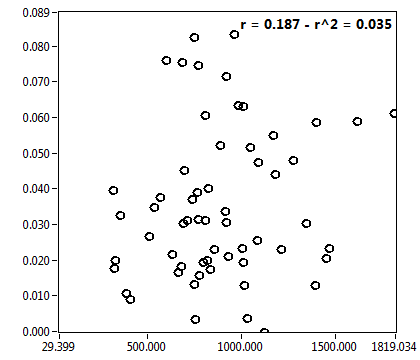
![c_corr [nM] - Ch1 - Cyto : ave = 288.351 - sd = 476.124](2093_c_corr_1c.png)
![Int_corr [kHz] - Ch1 - Cyto : ave = 28.433 - sd = 84.243](2093_Int_corr_1c.png)
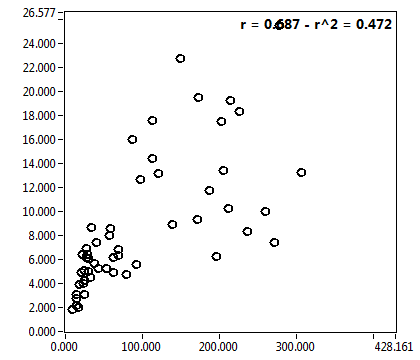
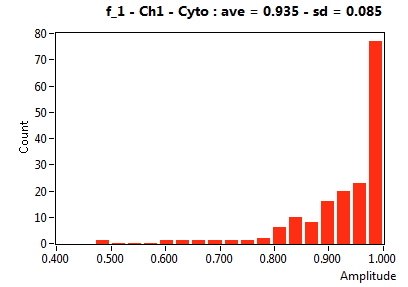
![D_1 [mum^2/s] - Ch1 - Cyto : ave = 38.512 - sd = 28.814](2093_D_1_1c.png)
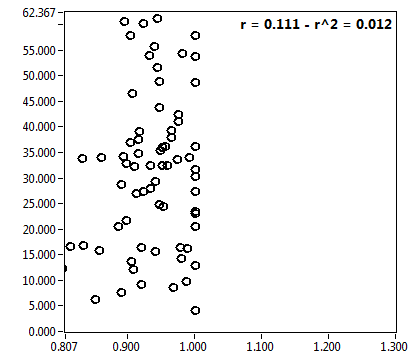
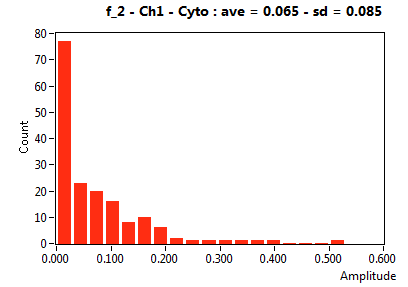
![D_2 [mum^2/s] - Ch1 - Cyto : ave = 1.005 - sd = 0.593](2093_D_2_1c.png)
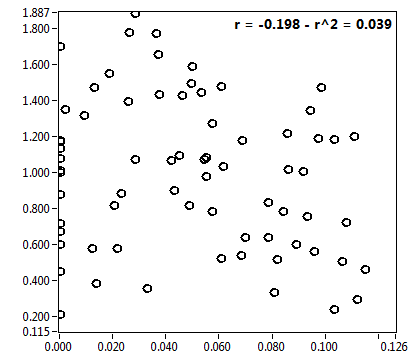
![CPM_corr [kHz] - Ch1 - Cyto : ave = 0.891 - sd = 1.015](2093_CPM_corr_1c.png)
![D_m [mum^2/s] - Ch1 - Cyto : ave = 36.804 - sd = 28.487](2093_D_m_1c.png)