 |  |
| 1 | 20110914\U03--V00--2176 |
|---|---|
| 2 | 20110920\U03--V00--2176 |
| 3 | 20110922\U02--V01--2176 |
| 4 | 20111123\U01--V01--2176 |
| 5 | 20120116\U02--V00--2176 |
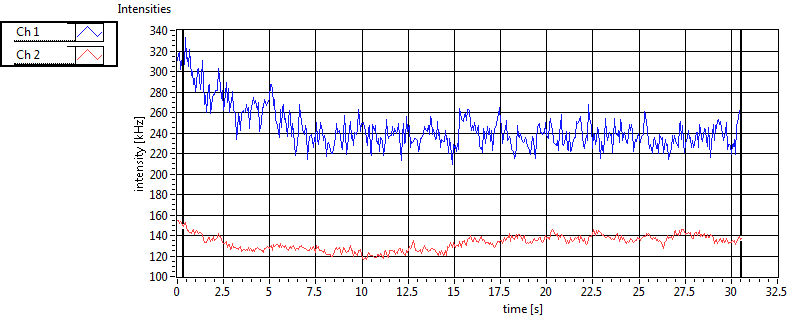
| Ch1 ave Nuc |
Ch1 sd Nuc |
Ch2 ave Nuc |
Ch2 sd Nuc |
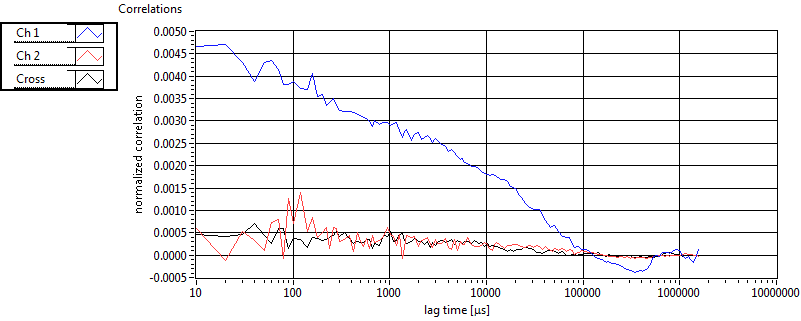
Cross ave Nuc |
Cross sd Nuc |
Ch1 ave Cyto |
Ch1 sd Cyto |
Ch2 ave Cyto |
Ch2 sd Cyto |
Cross ave Cyto |
Cross sd Cyto |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 1.461E+2 | 8.998E+1 | 1.032E+3 | 5.200E+2 | 2.040E+1 | 2.400E+1 | 2.023E+1 | 1.902E+1 | 2.739E+2 | 2.638E+2 | 1.331E+1 | 1.890E+1 |
| N_corr | 1.682E+2 | 1.142E+2 | 1.358E+3 | 7.936E+2 | 2.780E+1 | 3.895E+1 | 2.192E+1 | 2.177E+1 | 3.613E+2 | 4.536E+2 | 1.573E+1 | 2.173E+1 |
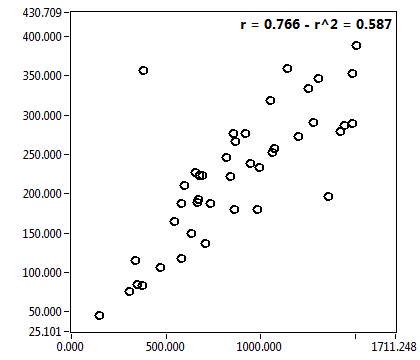
| c [nM] | 9.084E+2 | 5.596E+2 | 4.786E+3 | 2.412E+3 | 1.049E+2 | 1.234E+2 | 1.258E+2 | 1.183E+2 | 1.270E+3 | 1.223E+3 | 6.841E+1 | 9.717E+1 |
| c_corr [nM] | 1.046E+3 | 7.100E+2 | 6.299E+3 | 3.681E+3 | 1.429E+2 | 2.002E+2 | 1.363E+2 | 1.354E+2 | 1.676E+3 | 2.104E+3 | 8.085E+1 | 1.117E+2 |
| ratioG | 1.301E-1 | 8.594E-2 | 2.982E-1 | 2.094E-1 | ||||||||
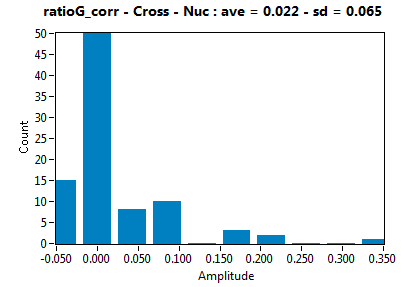
| ratioG_corr | 2.204E-2 | 6.519E-2 | 1.125E-1 | 9.090E-2 | ||||||||
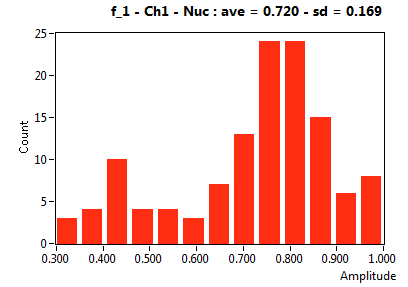
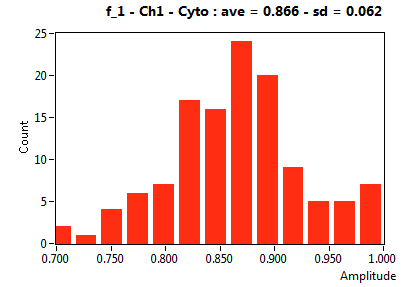
| f_1 | 7.199E-1 | 1.691E-1 | 4.537E-1 | 2.028E-1 | 9.494E-1 | 1.083E-1 | 8.661E-1 | 6.208E-2 | 8.328E-1 | 2.950E-1 | 9.696E-1 | 4.531E-2 |
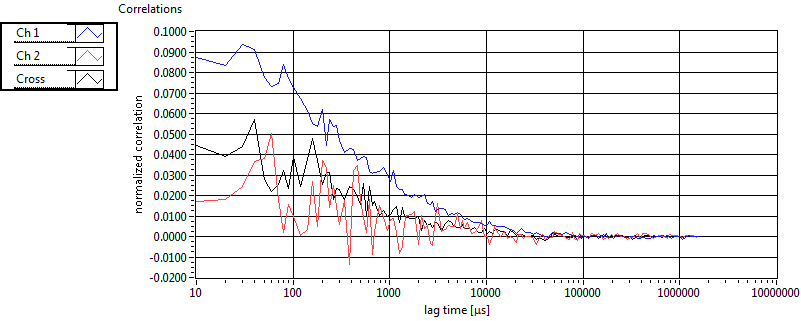
| tauD_1 [ms] | 2.560E+0 | 3.478E+0 | 1.839E+0 | 2.849E-2 | 2.071E+0 | 5.336E+0 | 6.972E-1 | 5.950E-1 | 1.776E+0 | 7.994E-1 | 1.211E+0 | 1.143E+0 |
| D_1 [mum^2/s] | 6.549E+0 | 4.817E+0 | 7.005E+0 | 1.096E-1 | 2.442E+1 | 3.128E+1 | 2.197E+1 | 1.128E+1 | 1.790E+1 | 3.222E+1 | 3.715E+1 | 3.691E+1 |
| alpha_1 | 9.805E-1 | 1.150E-1 | 9.203E-1 | 7.543E-4 | 7.372E-1 | 2.215E-1 | 9.955E-1 | 1.214E-1 | 1.163E+0 | 7.970E-1 | 9.507E-1 | 2.190E-1 |
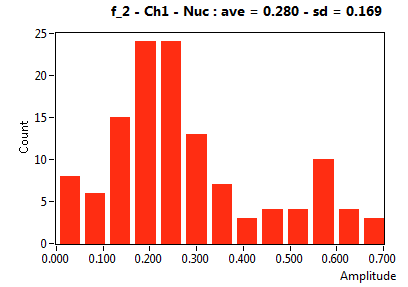
| f_2 | 2.801E-1 | 1.691E-1 | 5.463E-1 | 2.028E-1 | 5.056E-2 | 1.083E-1 | 1.339E-1 | 6.208E-2 | 1.672E-1 | 2.950E-1 | 3.043E-2 | 4.531E-2 |
| tauD_2 [ms] | 3.618E+1 | 3.461E+1 | 8.066E+1 | 9.920E+1 | 9.435E+1 | 1.048E+2 | 1.782E+1 | 1.308E+1 | 4.123E+1 | 5.708E+1 | 3.417E+1 | 2.356E+1 |
| D_2 [mum^2/s] | 3.782E-1 | 1.410E-1 | 4.962E-1 | 6.045E-1 | 1.987E-1 | 1.390E-1 | 8.444E-1 | 4.455E-1 | 1.359E+0 | 1.083E+0 | 6.435E-1 | 4.783E-1 |
| alpha_2 | 1.547E+0 | 3.270E-1 | 1.259E+0 | 3.379E-1 | 1.192E+0 | 2.679E-1 | 1.132E+0 | 2.143E-1 | 1.152E+0 | 2.401E-1 | 1.099E+0 | 2.114E-1 |
| tauD_m [ms] | 1.168E+1 | 9.090E+0 | 4.612E+1 | 6.605E+1 | 8.865E+0 | 3.229E+1 | 3.386E+0 | 3.419E+0 | 1.131E+1 | 2.155E+1 | 2.370E+0 | 2.299E+0 |
| D_m [mum^2/s] | 4.501E+0 | 2.456E+0 | 3.499E+0 | 1.216E+0 | 2.253E+1 | 2.828E+1 | 1.941E+1 | 1.058E+1 | 1.694E+1 | 3.258E+1 | 3.617E+1 | 3.624E+1 |
| R [1/s] | 1.741E+1 | 1.717E+1 | 2.252E+1 | 3.395E+1 | ||||||||
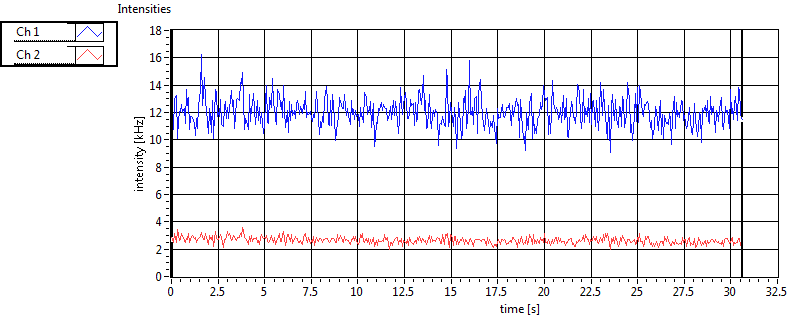
| CPM [kHz] | 1.623E+0 | 7.627E-1 | 3.385E-1 | 3.525E-1 | 1.117E+0 | 4.692E-1 | 2.793E-1 | 1.971E-1 | ||||
| CPM_corr [kHz] | 1.641E+0 | 7.671E-1 | 4.154E-1 | 4.347E-1 | 1.290E+0 | 5.070E-1 | 4.430E-1 | 2.089E-1 | ||||
| Int [kHz] | 2.335E+2 | 1.314E+2 | 1.777E+2 | 5.808E+1 | 2.332E+1 | 2.769E+1 | 6.317E+1 | 5.614E+1 | ||||
| Int_corr [kHz] | 2.687E+2 | 1.624E+2 | 2.792E+2 | 1.054E+2 | 2.746E+1 | 3.190E+1 | 9.779E+1 | 9.835E+1 | ||||
| # samples | 125 | 125 | 99 | 99 | 89 | 89 | 123 | 123 | 19 | 19 | 16 | 16 |
| # cells | 53 |
![c_corr [nM] - Ch1 - Nuc : ave = 1046.170 - sd = 709.968](2176_c_corr_1n.png)
![Int_corr [kHz] - Ch1 - Nuc : ave = 268.661 - sd = 162.373](2176_Int_corr_1n.png)
![D_1 [mum^2/s] - Ch1 - Nuc : ave = 6.549 - sd = 4.817](2176_D_1_1n.png)
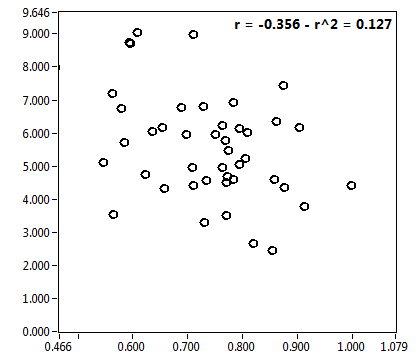
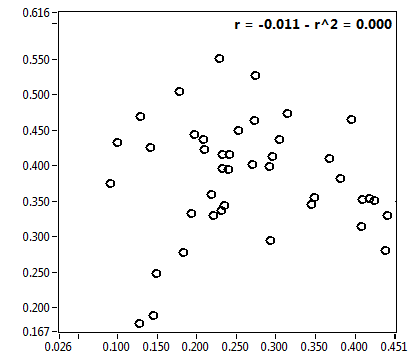
![D_2 [mum^2/s] - Ch1 - Nuc : ave = 0.378 - sd = 0.141](2176_D_2_1n.png)
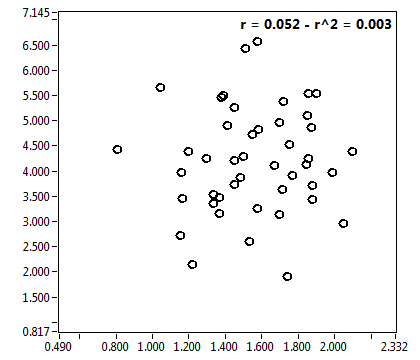
![CPM_corr [kHz] - Ch1 - Nuc : ave = 1.641 - sd = 0.767](2176_CPM_corr_1n.png)
![D_m [mum^2/s] - Ch1 - Nuc : ave = 4.501 - sd = 2.456](2176_D_m_1n.png)
![c_corr [nM] - Ch1 - Nuc : ave = 1046.170 - sd = 709.968](2176_c_corr_1n.png)

![c_corr [nM] - Ch1 - Cyto : ave = 136.346 - sd = 135.366](2176_c_corr_1c.png)
![Int_corr [kHz] - Ch1 - Cyto : ave = 27.460 - sd = 31.903](2176_Int_corr_1c.png)
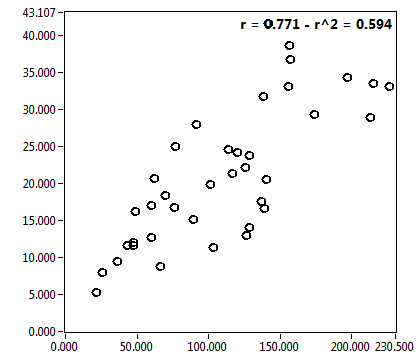
![D_1 [mum^2/s] - Ch1 - Cyto : ave = 21.974 - sd = 11.280](2176_D_1_1c.png)
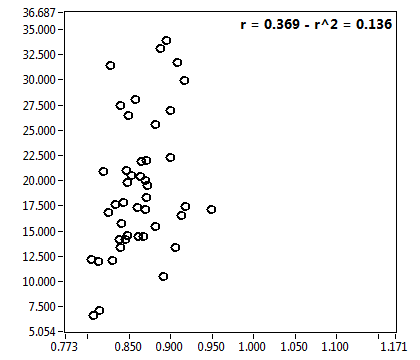
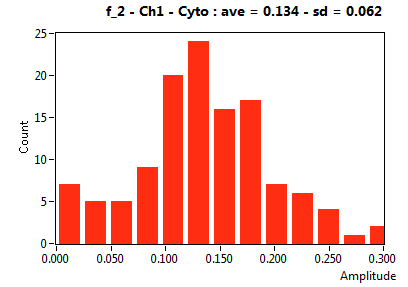
![D_2 [mum^2/s] - Ch1 - Cyto : ave = 0.844 - sd = 0.445](2176_D_2_1c.png)
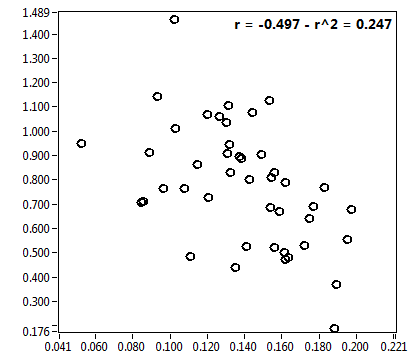
![CPM_corr [kHz] - Ch1 - Cyto : ave = 1.290 - sd = 0.507](2176_CPM_corr_1c.png)
![D_m [mum^2/s] - Ch1 - Cyto : ave = 19.406 - sd = 10.580](2176_D_m_1c.png)
