 |  |
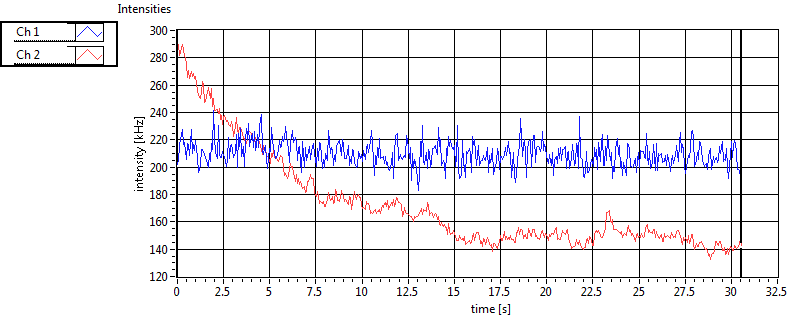
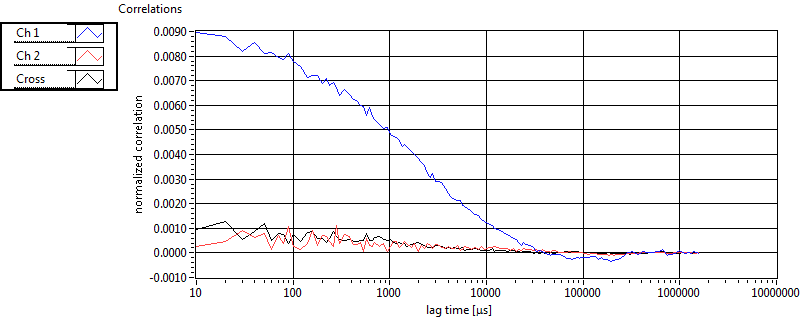
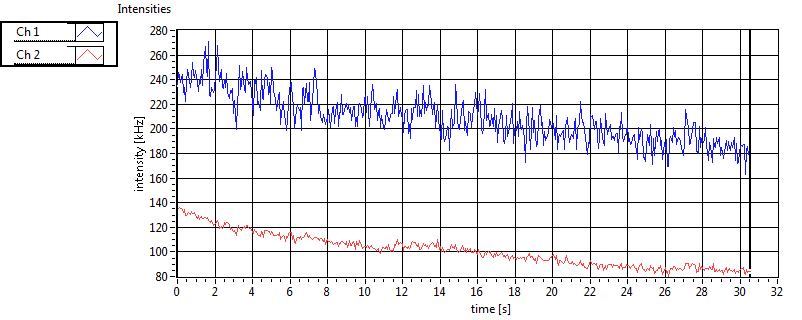
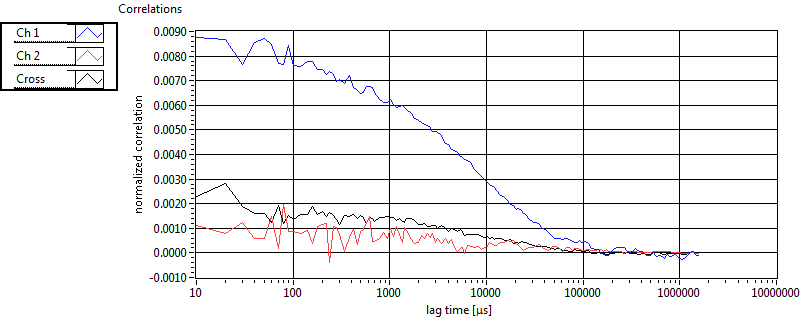
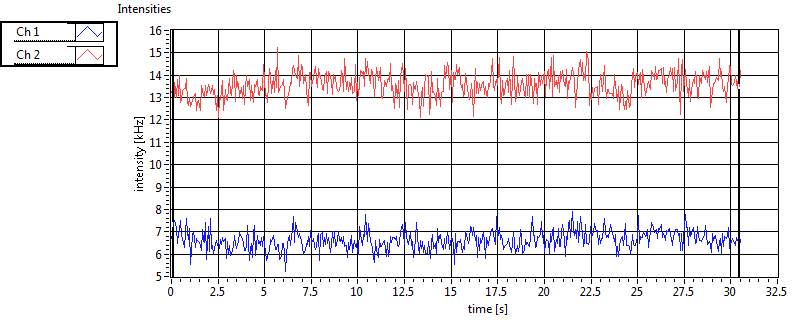
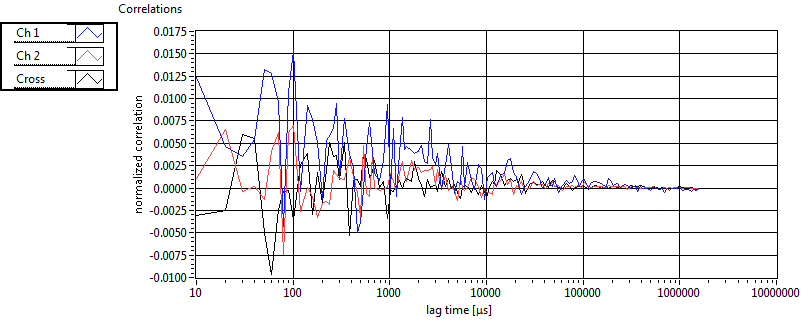
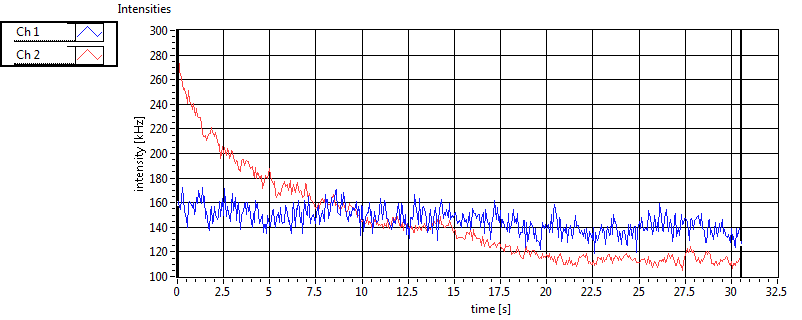
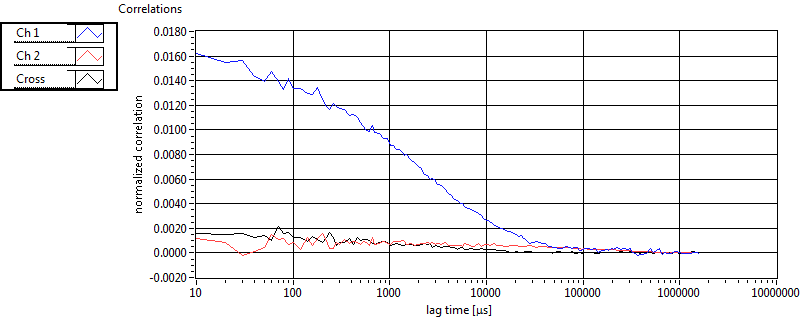
| 1 | 20110909\U02--V00--2432 |
|---|---|
| 2 | 20110923\U02--V00--2432 |
| 3 | 20110927\U02--V00--2432 |
| 4 | 20110928\U02--V00--2432 |
| Ch1 ave Nuc |
Ch1 sd Nuc |
Ch2 ave Nuc |
Ch2 sd Nuc |
Cross ave Nuc |
Cross sd Nuc |
Ch1 ave Cyto |
Ch1 sd Cyto |
Ch2 ave Cyto |
Ch2 sd Cyto |
Cross ave Cyto |
Cross sd Cyto |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 1.907E+2 | 2.025E+2 | 1.032E+3 | 3.679E+2 | 9.284E+1 | 1.266E+2 | 5.055E+1 | 5.914E+1 | 5.388E+2 | 3.406E+2 | 2.867E+1 | 3.926E+1 |
| N_corr | 2.100E+2 | 2.127E+2 | 1.099E+3 | 6.189E+2 | 1.308E+2 | 1.675E+2 | 4.422E+1 | 5.494E+1 | 6.620E+2 | 4.625E+2 | 2.648E+1 | 3.580E+1 |
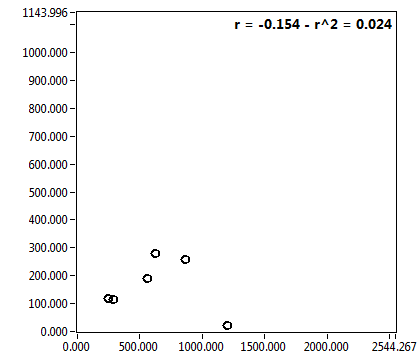
| c [nM] | 1.186E+3 | 1.260E+3 | 4.788E+3 | 1.706E+3 | 4.773E+2 | 6.509E+2 | 3.144E+2 | 3.678E+2 | 2.499E+3 | 1.580E+3 | 1.474E+2 | 2.019E+2 |
| c_corr [nM] | 1.306E+3 | 1.323E+3 | 5.096E+3 | 2.871E+3 | 6.727E+2 | 8.609E+2 | 2.750E+2 | 3.417E+2 | 3.071E+3 | 2.145E+3 | 1.361E+2 | 1.841E+2 |
| ratioG | 2.376E-1 | 1.922E-1 | 2.236E-1 | 1.636E-1 | ||||||||
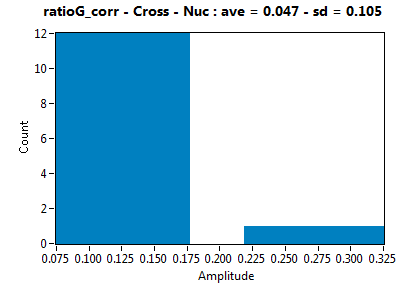
| ratioG_corr | 4.743E-2 | 1.051E-1 | 1.393E-1 | 1.479E-1 | ||||||||
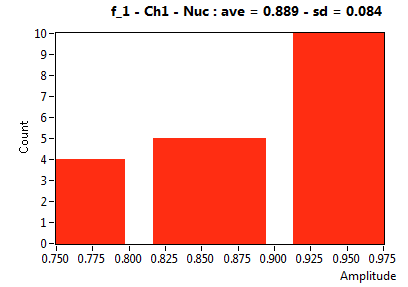
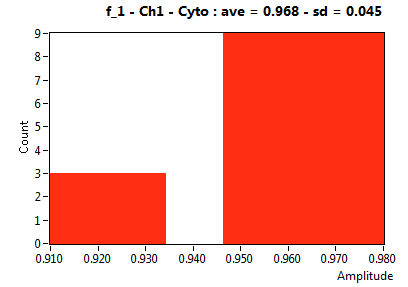
| f_1 | 8.891E-1 | 8.355E-2 | 5.558E-1 | 1.900E-1 | 9.193E-1 | 9.038E-2 | 9.676E-1 | 4.539E-2 | 7.285E-1 | 3.450E-1 | 9.432E-1 | 3.625E-2 |
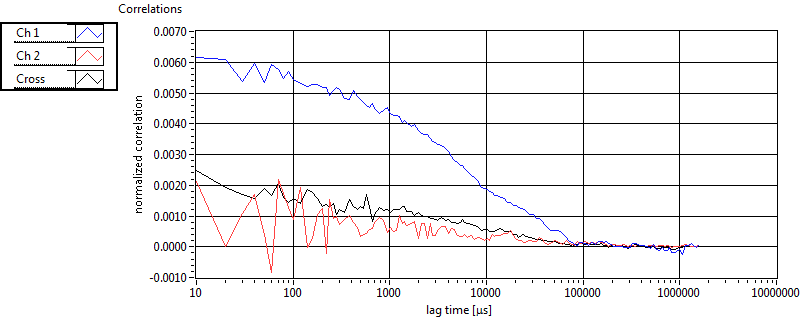
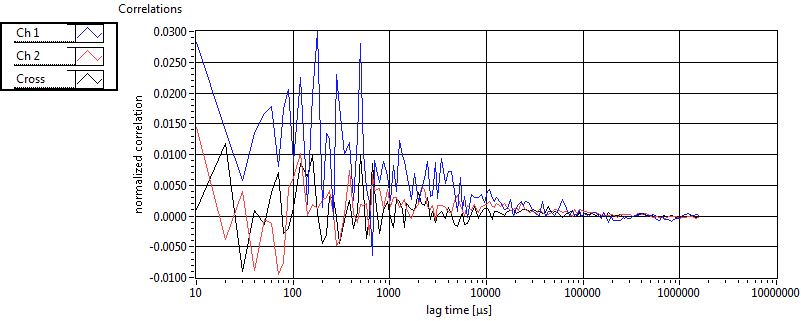
| tauD_1 [ms] | 2.918E+0 | 1.459E+0 | 1.823E+0 | 2.919E-2 | 1.752E+0 | 9.985E-1 | 1.705E+0 | 7.624E-1 | 2.368E+0 | 9.249E-1 | 1.585E+0 | 5.949E-1 |
| D_1 [mum^2/s] | 4.333E+0 | 1.483E+0 | 7.068E+0 | 1.123E-1 | 9.863E+0 | 7.813E+0 | 8.815E+0 | 7.067E+0 | 6.033E+0 | 1.528E+0 | 8.796E+0 | 3.292E+0 |
| alpha_1 | 1.068E+0 | 7.379E-2 | 9.200E-1 | 0.000E+0 | 8.233E-1 | 1.251E-1 | 1.009E+0 | 1.632E-1 | 1.032E+0 | 1.372E-1 | 1.073E+0 | 2.197E-1 |
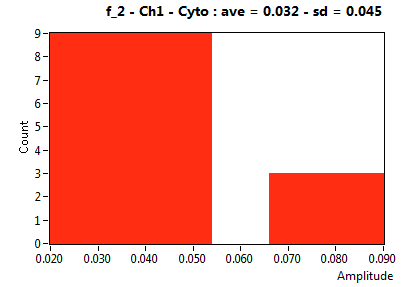
| f_2 | 1.109E-1 | 8.355E-2 | 4.442E-1 | 1.900E-1 | 8.067E-2 | 9.038E-2 | 3.240E-2 | 4.539E-2 | 2.715E-1 | 3.450E-1 | 5.676E-2 | 3.625E-2 |
| tauD_2 [ms] | 2.735E+1 | 1.239E+1 | 8.596E+1 | 5.462E+1 | 4.285E+1 | 1.743E+1 | 2.776E+1 | 9.730E+0 | 1.010E+2 | 1.329E+2 | 5.259E+1 | 1.554E+1 |
| D_2 [mum^2/s] | 4.645E-1 | 1.835E-1 | 2.651E-1 | 2.218E-1 | 3.303E-1 | 1.399E-1 | 4.287E-1 | 1.380E-1 | 1.118E+0 | 1.193E+0 | 2.503E-1 | 7.677E-2 |
| alpha_2 | 1.291E+0 | 2.470E-1 | 1.323E+0 | 2.796E-1 | 1.333E+0 | 3.218E-1 | 9.780E-1 | 1.684E-1 | 9.736E-1 | 2.710E-1 | 9.777E-1 | 2.795E-1 |
| tauD_m [ms] | 5.417E+0 | 2.632E+0 | 4.249E+1 | 3.498E+1 | 4.214E+0 | 2.649E+0 | 2.378E+0 | 1.389E+0 | 4.690E+1 | 5.853E+1 | 4.263E+0 | 2.673E+0 |
| D_m [mum^2/s] | 3.897E+0 | 1.347E+0 | 4.051E+0 | 1.354E+0 | 8.844E+0 | 6.390E+0 | 8.575E+0 | 6.986E+0 | 4.146E+0 | 2.053E+0 | 8.402E+0 | 3.363E+0 |
| R [1/s] | 3.610E+1 | 2.766E+1 | 1.706E+1 | 1.828E+1 | ||||||||
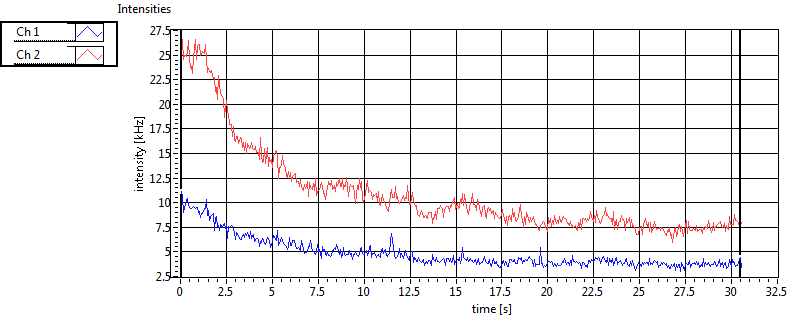
| CPM [kHz] | 2.208E+0 | 6.465E-1 | 2.999E-1 | 1.872E-1 | 6.702E-1 | 7.036E-1 | 3.391E-1 | 1.967E-1 | ||||
| CPM_corr [kHz] | 2.231E+0 | 6.532E-1 | 4.610E-1 | 2.402E-1 | 8.689E-1 | 7.312E-1 | 3.849E-1 | 2.125E-1 | ||||
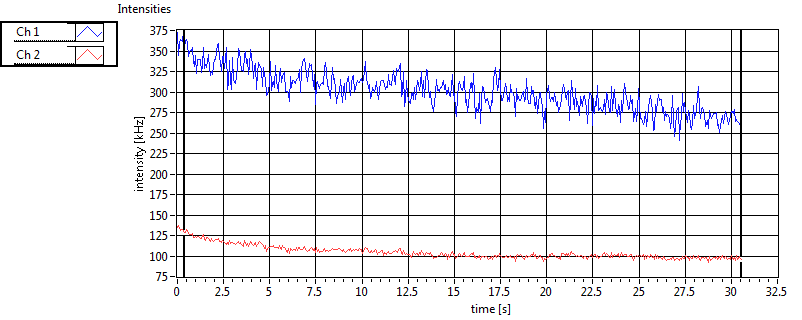
| Int [kHz] | 4.201E+2 | 4.689E+2 | 1.766E+2 | 5.436E+1 | 2.811E+1 | 4.222E+1 | 8.809E+1 | 4.337E+1 | ||||
| Int_corr [kHz] | 4.752E+2 | 5.048E+2 | 2.722E+2 | 7.180E+1 | 2.863E+1 | 4.351E+1 | 1.237E+2 | 6.177E+1 | ||||
| # samples | 19 | 19 | 13 | 13 | 13 | 13 | 12 | 12 | 5 | 5 | 4 | 4 |
| # cells | 8 |
![c_corr [nM] - Ch1 - Nuc : ave = 1306.243 - sd = 1323.131](2432_c_corr_1n.png)
![Int_corr [kHz] - Ch1 - Nuc : ave = 475.234 - sd = 504.816](2432_Int_corr_1n.png)
![D_1 [mum^2/s] - Ch1 - Nuc : ave = 4.333 - sd = 1.483](2432_D_1_1n.png)
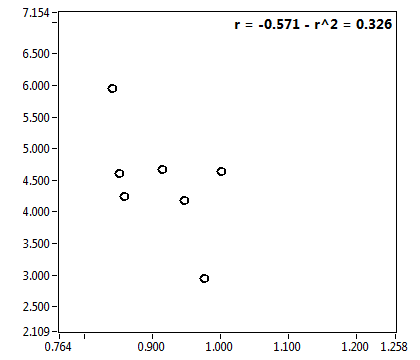
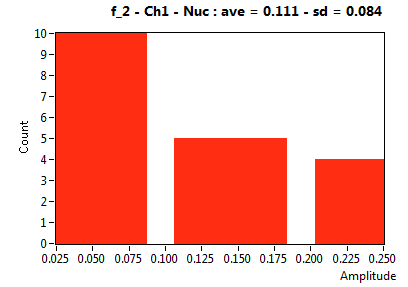
![D_2 [mum^2/s] - Ch1 - Nuc : ave = 0.465 - sd = 0.183](2432_D_2_1n.png)
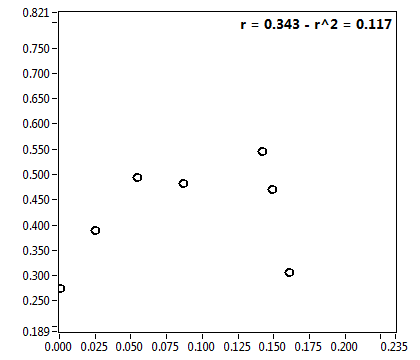
![CPM_corr [kHz] - Ch1 - Nuc : ave = 2.231 - sd = 0.653](2432_CPM_corr_1n.png)
![D_m [mum^2/s] - Ch1 - Nuc : ave = 3.897 - sd = 1.347](2432_D_m_1n.png)
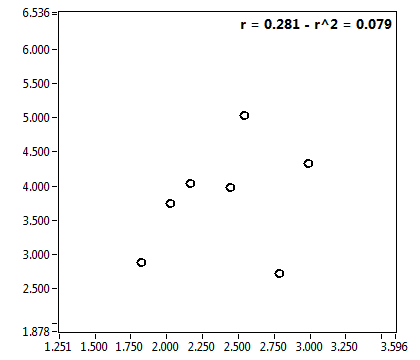
![c_corr [nM] - Ch1 - Nuc : ave = 1306.243 - sd = 1323.131](2432_c_corr_1n.png)

![c_corr [nM] - Ch1 - Cyto : ave = 275.012 - sd = 341.697](2432_c_corr_1c.png)
![Int_corr [kHz] - Ch1 - Cyto : ave = 28.630 - sd = 43.514](2432_Int_corr_1c.png)
![D_1 [mum^2/s] - Ch1 - Cyto : ave = 8.815 - sd = 7.067](2432_D_1_1c.png)
![D_2 [mum^2/s] - Ch1 - Cyto : ave = 0.429 - sd = 0.138](2432_D_2_1c.png)
![CPM_corr [kHz] - Ch1 - Cyto : ave = 0.869 - sd = 0.731](2432_CPM_corr_1c.png)
![D_m [mum^2/s] - Ch1 - Cyto : ave = 8.575 - sd = 6.986](2432_D_m_1c.png)
