 |  |
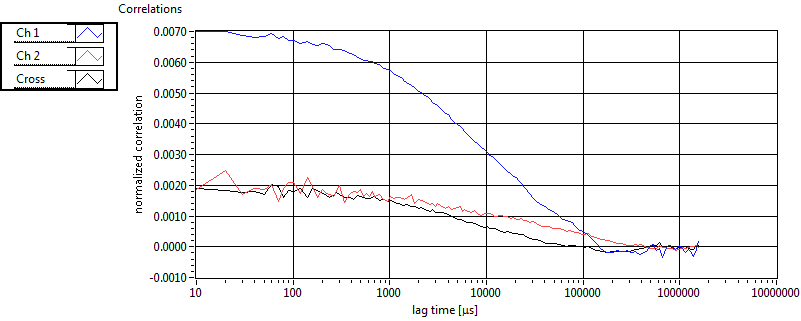
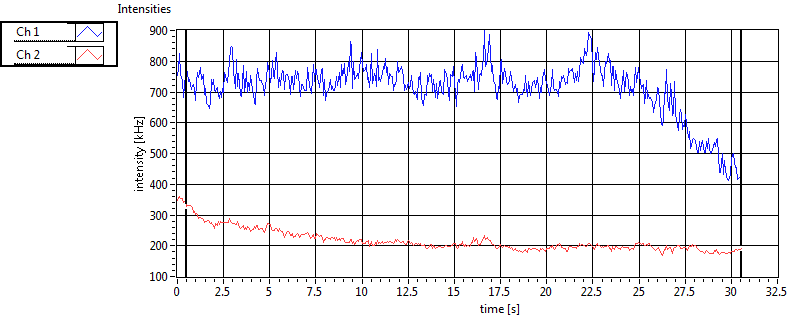
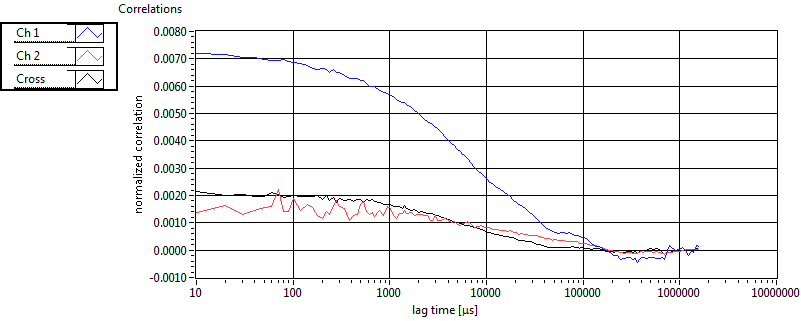
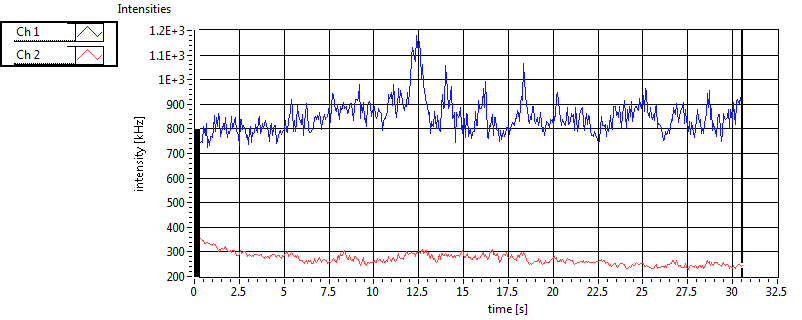
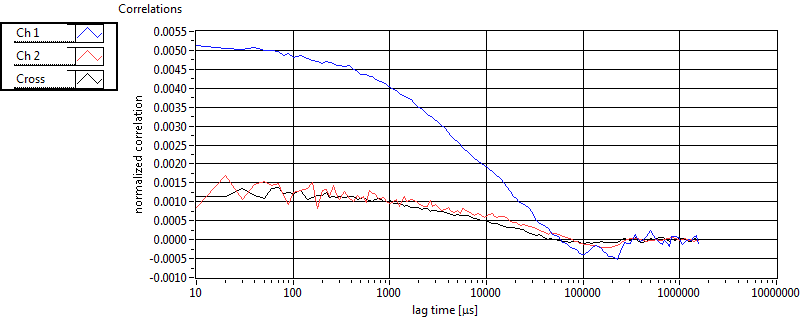
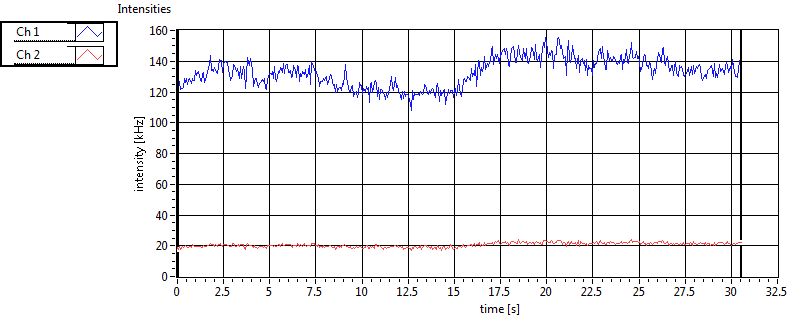
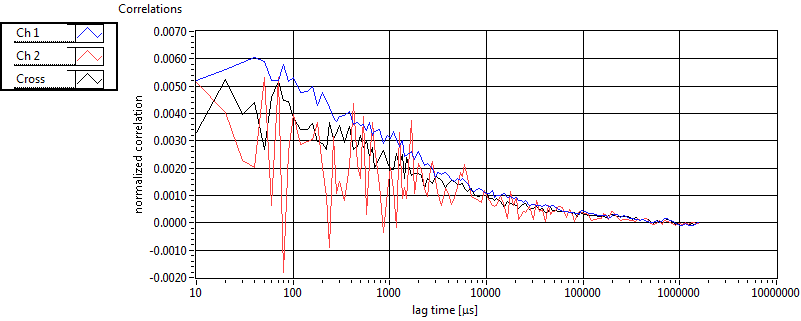
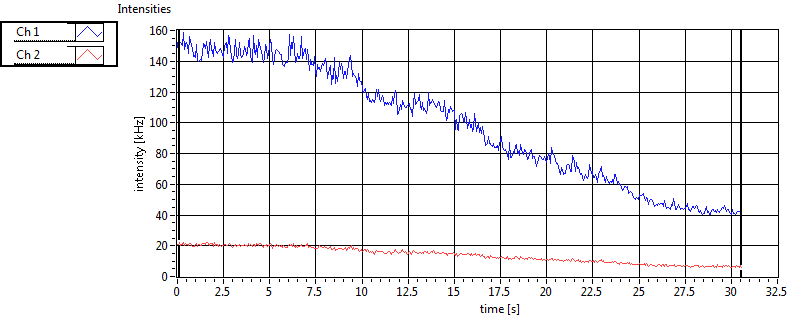
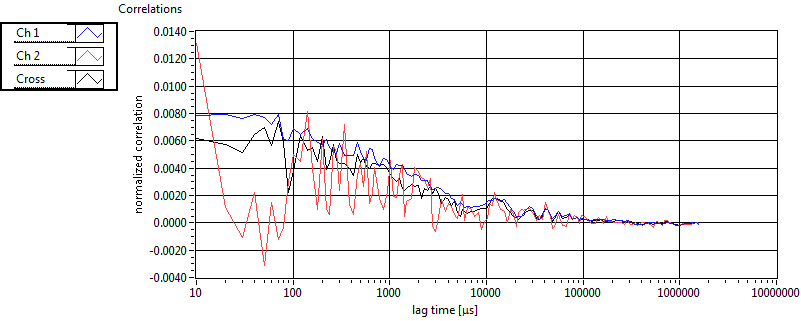
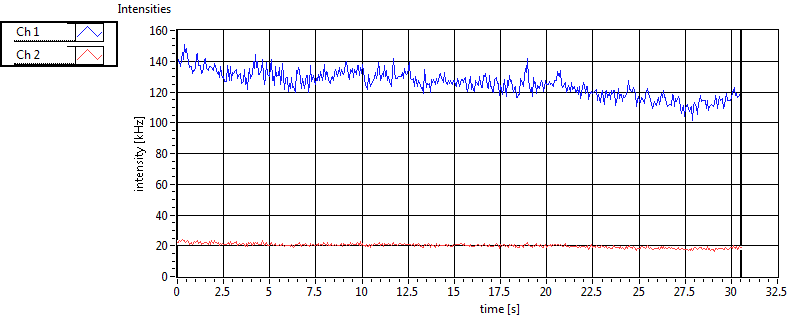
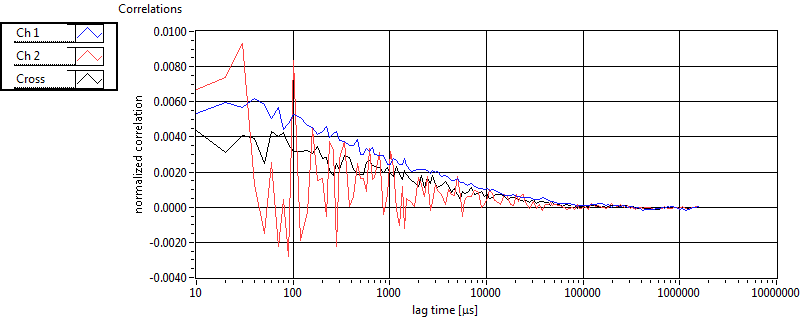
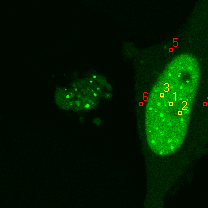
| 1 | 20130109\U05--V00--2960hi |
|---|---|
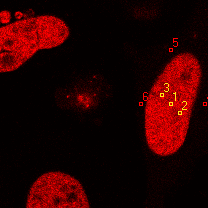
| 2 | 20130111\U01--V01--2960hi |
| Ch1 ave Nuc |
Ch1 sd Nuc |
Ch2 ave Nuc |
Ch2 sd Nuc |
Cross ave Nuc |
Cross sd Nuc |
Ch1 ave Cyto |
Ch1 sd Cyto |
Ch2 ave Cyto |
Ch2 sd Cyto |
Cross ave Cyto |
Cross sd Cyto |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 1.891E+2 | 1.225E+2 | 8.633E+2 | 4.094E+2 | 3.562E+1 | 3.261E+1 | 1.428E+2 | 1.437E+2 | 5.325E+2 | 3.145E+2 | 6.469E+1 | 6.680E+1 |
| N_corr | 1.908E+2 | 1.217E+2 | 1.214E+3 | 7.264E+2 | 3.885E+1 | 3.716E+1 | 1.516E+2 | 1.571E+2 | 9.094E+2 | 5.907E+2 | 7.840E+1 | 7.058E+1 |
| c [nM] | 1.176E+3 | 7.617E+2 | 4.004E+3 | 1.899E+3 | 1.831E+2 | 1.677E+2 | 8.881E+2 | 8.936E+2 | 2.470E+3 | 1.459E+3 | 3.326E+2 | 3.434E+2 |
| c_corr [nM] | 1.187E+3 | 7.566E+2 | 5.629E+3 | 3.369E+3 | 1.997E+2 | 1.910E+2 | 9.426E+2 | 9.773E+2 | 4.218E+3 | 2.740E+3 | 4.030E+2 | 3.629E+2 |
| ratioG | 1.454E-1 | 8.751E-2 | 3.133E-1 | 2.359E-1 | ||||||||
| ratioG_corr | 5.083E-3 | 5.534E-2 | 1.182E-1 | 9.826E-2 | ||||||||
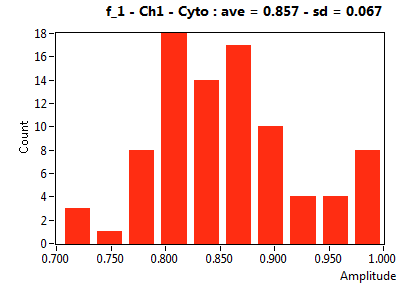
| f_1 | 8.822E-1 | 1.031E-1 | 3.832E-1 | 2.418E-1 | 9.584E-1 | 1.131E-1 | 8.569E-1 | 6.679E-2 | 6.564E-1 | 2.904E-1 | 9.183E-1 | 9.089E-2 |
| tauD_1 [ms] | 2.015E+0 | 2.219E+0 | 1.860E+0 | 4.441E-16 | 2.162E+0 | 2.332E+0 | 1.093E+0 | 7.491E-1 | 1.860E+0 | 0.000E+0 | 1.019E+0 | 6.590E-1 |
| D_1 [mum^2/s] | 9.997E+0 | 8.320E+0 | 6.926E+0 | 8.882E-16 | 1.816E+1 | 1.737E+1 | 1.288E+1 | 6.609E+0 | 6.926E+0 | 0.000E+0 | 1.483E+1 | 5.637E+0 |
| alpha_1 | 9.985E-1 | 1.045E-1 | 9.200E-1 | 1.110E-16 | 9.727E-1 | 2.102E-1 | 1.021E+0 | 1.161E-1 | 9.200E-1 | 0.000E+0 | 9.763E-1 | 1.924E-1 |
| f_2 | 1.178E-1 | 1.031E-1 | 6.168E-1 | 2.418E-1 | 4.155E-2 | 1.131E-1 | 1.431E-1 | 6.679E-2 | 3.436E-1 | 2.904E-1 | 8.169E-2 | 9.089E-2 |
| tauD_2 [ms] | 3.330E+1 | 3.991E+1 | 5.443E+1 | 6.699E+1 | 1.005E+2 | 8.375E+1 | 3.682E+1 | 3.328E+1 | 2.683E+1 | 1.890E+1 | 4.167E+1 | 1.225E+1 |
| D_2 [mum^2/s] | 5.024E-1 | 2.497E-1 | 7.567E-1 | 9.084E-1 | 1.628E-1 | 7.280E-2 | 4.218E-1 | 2.200E-1 | 9.689E-1 | 9.766E-1 | 3.179E-1 | 1.077E-1 |
| alpha_2 | 1.326E+0 | 3.196E-1 | 1.331E+0 | 3.435E-1 | 1.132E+0 | 1.635E-1 | 1.132E+0 | 1.933E-1 | 1.113E+0 | 9.159E-2 | 1.318E+0 | 3.702E-1 |
| tauD_m [ms] | 5.887E+0 | 7.629E+0 | 3.394E+1 | 5.043E+1 | 5.203E+0 | 8.686E+0 | 6.095E+0 | 5.671E+0 | 1.026E+1 | 1.527E+1 | 4.085E+0 | 4.453E+0 |
| D_m [mum^2/s] | 8.994E+0 | 7.901E+0 | 3.210E+0 | 1.358E+0 | 1.786E+1 | 1.738E+1 | 1.106E+1 | 5.823E+0 | 4.946E+0 | 1.728E+0 | 1.371E+1 | 5.314E+0 |
| R [1/s] | 1.116E+1 | 1.874E+1 | 9.285E+0 | 1.436E+1 | ||||||||
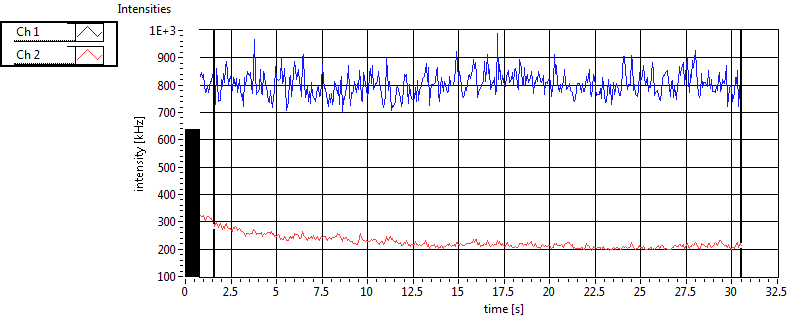
| CPM [kHz] | 9.306E-1 | 9.059E-1 | 3.673E-1 | 3.329E-1 | 6.684E-1 | 1.923E-1 | 2.709E-1 | 1.836E-1 | ||||
| CPM_corr [kHz] | 9.446E-1 | 9.051E-1 | 4.466E-1 | 4.092E-1 | 6.903E-1 | 1.918E-1 | 3.971E-1 | 2.079E-1 | ||||
| Int [kHz] | 1.934E+2 | 2.314E+2 | 1.767E+2 | 5.562E+1 | 9.229E+1 | 6.762E+1 | 1.076E+2 | 5.991E+1 | ||||
| Int_corr [kHz] | 1.986E+2 | 2.368E+2 | 2.849E+2 | 1.007E+2 | 1.008E+2 | 7.876E+1 | 2.415E+2 | 1.355E+2 | ||||
| # samples | 114 | 114 | 98 | 98 | 43 | 43 | 87 | 87 | 16 | 16 | 10 | 10 |
| # cells | 55 |
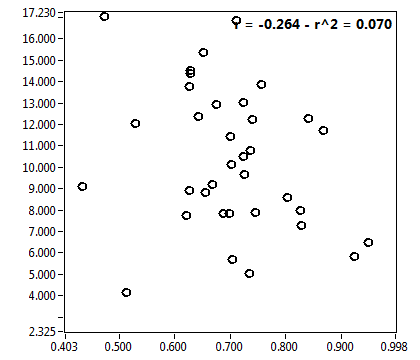
![c_corr [nM] - Ch1 - Nuc : ave = 1186.717 - sd = 756.575](2960hi_c_corr_1n.png)
![Int_corr [kHz] - Ch1 - Nuc : ave = 198.576 - sd = 236.785](2960hi_Int_corr_1n.png)
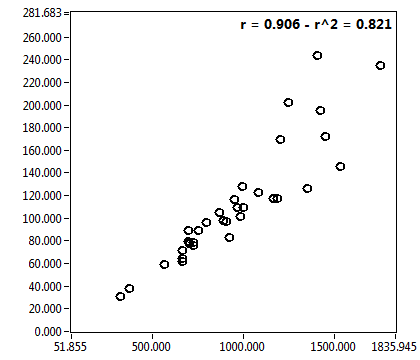
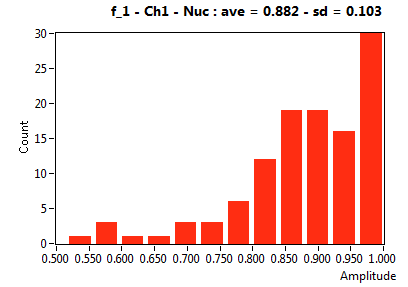
![D_1 [mum^2/s] - Ch1 - Nuc : ave = 9.997 - sd = 8.320](2960hi_D_1_1n.png)
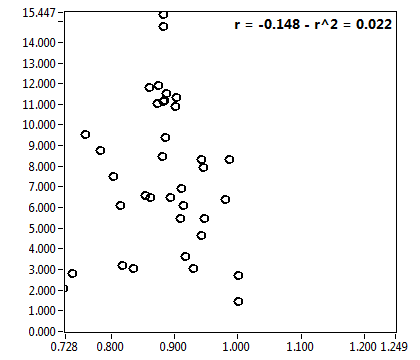
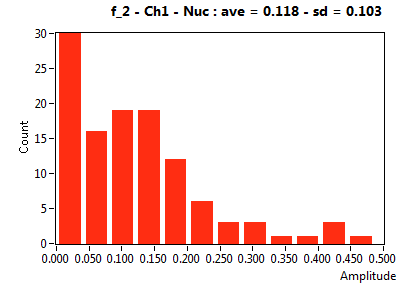
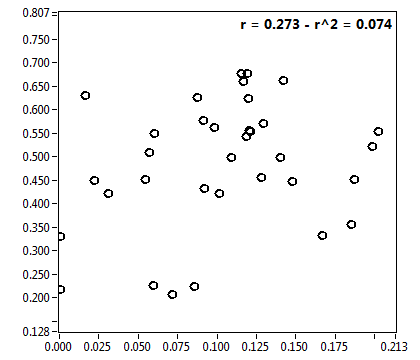
![D_2 [mum^2/s] - Ch1 - Nuc : ave = 0.502 - sd = 0.250](2960hi_D_2_1n.png)
![CPM_corr [kHz] - Ch1 - Nuc : ave = 0.945 - sd = 0.905](2960hi_CPM_corr_1n.png)
![D_m [mum^2/s] - Ch1 - Nuc : ave = 8.994 - sd = 7.901](2960hi_D_m_1n.png)
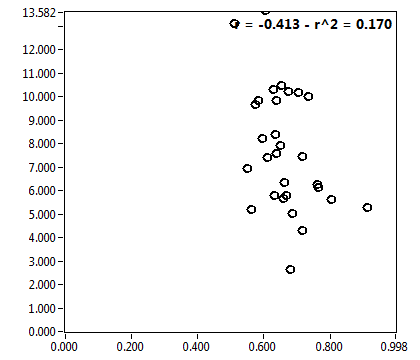
![c_corr [nM] - Ch1 - Nuc : ave = 1186.717 - sd = 756.575](2960hi_c_corr_1n.png)
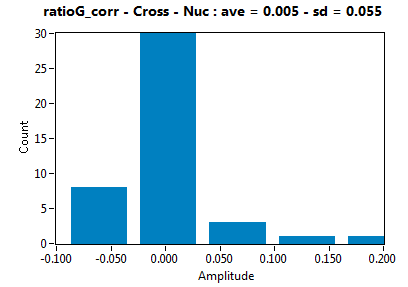
![c_corr [nM] - Ch1 - Cyto : ave = 942.567 - sd = 977.331](2960hi_c_corr_1c.png)
![Int_corr [kHz] - Ch1 - Cyto : ave = 100.839 - sd = 78.760](2960hi_Int_corr_1c.png)

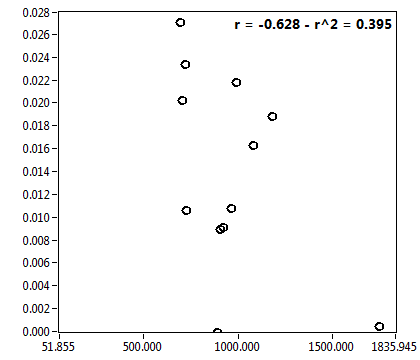
![D_1 [mum^2/s] - Ch1 - Cyto : ave = 12.881 - sd = 6.609](2960hi_D_1_1c.png)
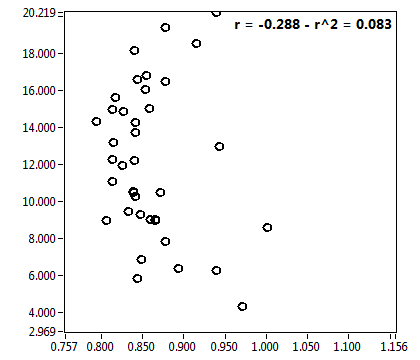
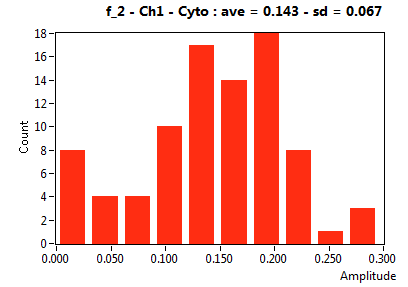
![D_2 [mum^2/s] - Ch1 - Cyto : ave = 0.422 - sd = 0.220](2960hi_D_2_1c.png)
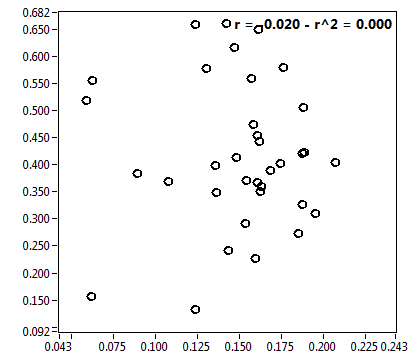
![CPM_corr [kHz] - Ch1 - Cyto : ave = 0.690 - sd = 0.192](2960hi_CPM_corr_1c.png)
![D_m [mum^2/s] - Ch1 - Cyto : ave = 11.059 - sd = 5.823](2960hi_D_m_1c.png)