 |  |
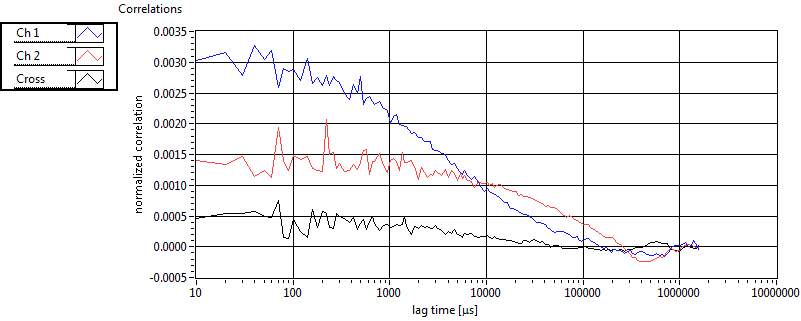
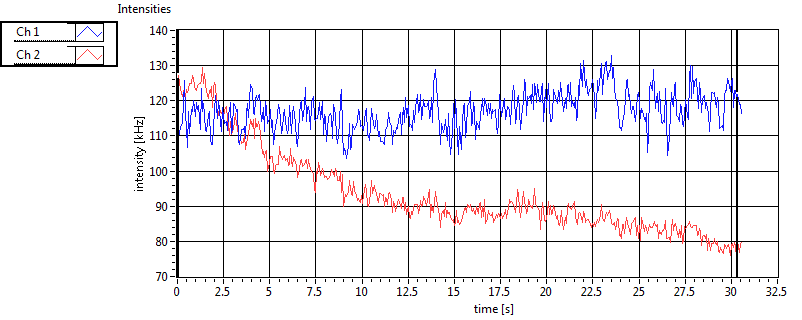
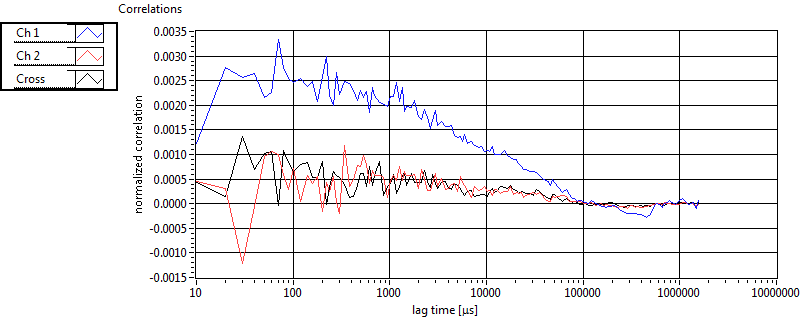
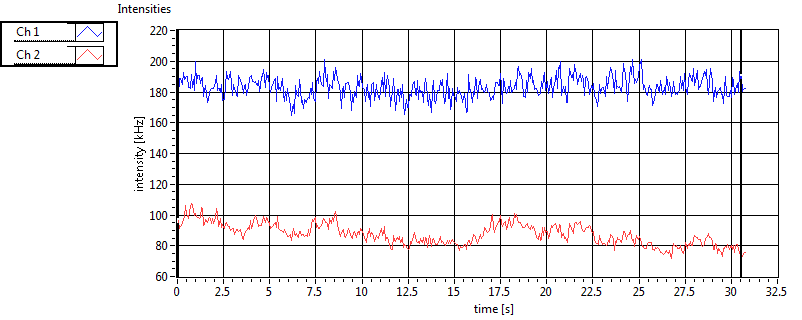
| 1 | 20130109\U01--V00--HP1hi |
|---|---|
| 2 | 20130110\U00--V01--HP1hi |
| 3 | 20130117\X00--Y00--HP1hi |
| 4 | 20130118\X00--Y00--HP1hi |
| 5 | 20130118\X01--Y00--HP1hi |
| 6 | 20130118\X02--Y00--HP1hi |
| 7 | 20130118\X03--Y00--HP1hi |
| 8 | 20130125\U01--V00--HP1hi |
| 9 | 20130125\U01--V01--HP1hi |
| 10 | 20130129\X00--Y00--HP1hi |
| 11 | 20130129\X01--Y00--HP1hi |
| 12 | 20130129\X02--Y00--HP1hi |
| 13 | 20130129\X03--Y00--HP1hi |
| 14 | 20130129\X05--Y00--HP1hi |
| 15 | 20130130\X00--Y00--HP1hi |
| 16 | 20130130\X01--Y00--HP1hi |
| 17 | 20130130\X03--Y00--HP1hi |
| 18 | 20130131\U00--V01--HP1hi |
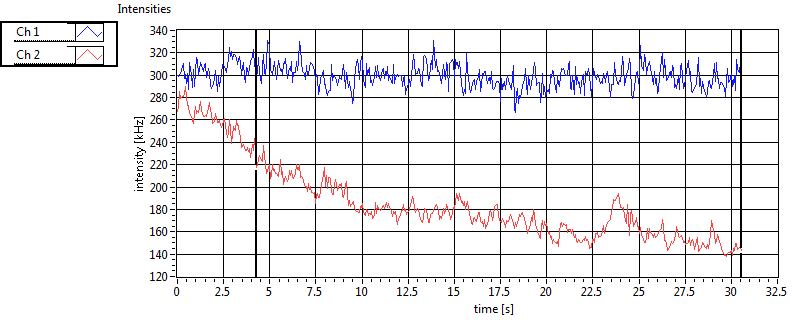
| Ch1 ave Nuc |
Ch1 sd Nuc |
Ch2 ave Nuc |
Ch2 sd Nuc |
Cross ave Nuc |
Cross sd Nuc |
Ch1 ave Cyto |
Ch1 sd Cyto |
Ch2 ave Cyto |
Ch2 sd Cyto |
Cross ave Cyto |
Cross sd Cyto |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 5.973E+2 | 3.563E+2 | 1.531E+3 | 7.982E+2 | 1.339E+2 | 1.043E+2 | 7.103E+1 | 1.130E+2 | 4.091E+2 | 2.769E+2 | 6.911E+1 | 4.669E+1 |
| N_corr | 6.016E+2 | 3.604E+2 | 1.617E+3 | 8.803E+2 | 1.425E+2 | 1.152E+2 | 6.843E+1 | 1.181E+2 | 4.368E+2 | 3.124E+2 | 7.791E+1 | 5.712E+1 |
| c [nM] | 3.715E+3 | 2.216E+3 | 7.101E+3 | 3.702E+3 | 6.882E+2 | 5.364E+2 | 4.418E+2 | 7.029E+2 | 1.898E+3 | 1.284E+3 | 3.553E+2 | 2.400E+2 |
| c_corr [nM] | 3.741E+3 | 2.241E+3 | 7.501E+3 | 4.083E+3 | 7.328E+2 | 5.920E+2 | 4.256E+2 | 7.343E+2 | 2.026E+3 | 1.449E+3 | 4.005E+2 | 2.936E+2 |
| ratioG | 2.479E-1 | 1.148E-1 | 3.881E-1 | 1.633E-1 | ||||||||
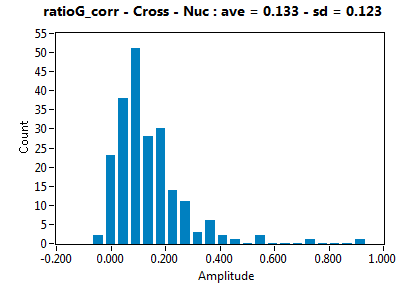
| ratioG_corr | 1.334E-1 | 1.233E-1 | 2.771E-1 | 1.871E-1 | ||||||||
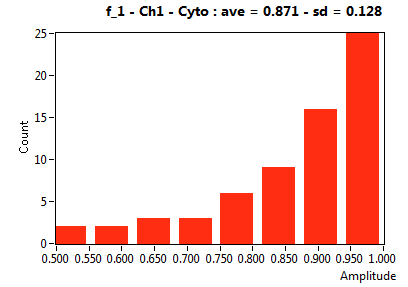
| f_1 | 8.593E-1 | 1.788E-1 | 2.601E-1 | 1.948E-1 | 7.723E-1 | 2.029E-1 | 8.711E-1 | 1.282E-1 | 5.188E-1 | 2.246E-1 | 7.278E-1 | 1.605E-1 |
| tauD_1 [ms] | 6.823E+0 | 4.343E+0 | 1.860E+0 | 8.882E-16 | 5.645E+0 | 9.893E+0 | 1.204E+0 | 1.471E+0 | 1.860E+0 | 0.000E+0 | 3.395E+0 | 4.437E+0 |
| D_1 [mum^2/s] | 2.470E+0 | 4.369E+0 | 6.926E+0 | 6.217E-15 | 9.342E+0 | 1.756E+1 | 2.601E+1 | 2.487E+1 | 6.926E+0 | 0.000E+0 | 6.202E+0 | 3.615E+0 |
| alpha_1 | 9.714E-1 | 1.300E-1 | 9.232E-1 | 2.378E-2 | 9.340E-1 | 2.376E-1 | 9.929E-1 | 1.926E-1 | 9.200E-1 | 0.000E+0 | 9.976E-1 | 1.813E-1 |
| f_2 | 1.407E-1 | 1.788E-1 | 7.399E-1 | 1.948E-1 | 2.277E-1 | 2.029E-1 | 1.289E-1 | 1.282E-1 | 4.812E-1 | 2.246E-1 | 2.722E-1 | 1.605E-1 |
| tauD_2 [ms] | 5.897E+1 | 3.106E+1 | 6.723E+1 | 3.300E+1 | 1.195E+2 | 7.564E+1 | 3.427E+1 | 4.643E+1 | 1.125E+2 | 7.318E+1 | 1.025E+2 | 7.603E+1 |
| D_2 [mum^2/s] | 2.174E-1 | 9.775E-2 | 2.529E-1 | 2.140E-1 | 1.366E-1 | 7.491E-2 | 6.722E-1 | 4.278E-1 | 1.821E-1 | 1.395E-1 | 1.621E-1 | 7.122E-2 |
| alpha_2 | 1.272E+0 | 2.858E-1 | 1.432E+0 | 3.187E-1 | 1.473E+0 | 3.995E-1 | 1.191E+0 | 2.605E-1 | 1.616E+0 | 3.615E-1 | 1.578E+0 | 4.038E-1 |
| tauD_m [ms] | 1.433E+1 | 1.122E+1 | 4.846E+1 | 2.419E+1 | 3.562E+1 | 4.425E+1 | 8.463E+0 | 1.856E+1 | 5.039E+1 | 4.044E+1 | 3.634E+1 | 4.272E+1 |
| D_m [mum^2/s] | 1.937E+0 | 2.887E+0 | 1.994E+0 | 1.285E+0 | 6.689E+0 | 1.166E+1 | 2.393E+1 | 2.443E+1 | 3.699E+0 | 1.458E+0 | 4.320E+0 | 2.241E+0 |
| R [1/s] | 5.590E+0 | 1.239E+1 | 3.976E+0 | 6.206E+0 | ||||||||
| CPM [kHz] | 4.720E-1 | 2.560E-1 | 3.367E-1 | 1.796E-1 | 4.204E-1 | 4.053E-1 | 4.867E-1 | 5.244E-1 | ||||
| CPM_corr [kHz] | 4.776E-1 | 2.549E-1 | 3.866E-1 | 2.137E-1 | 7.090E-1 | 6.595E-1 | 5.807E-1 | 5.526E-1 | ||||
| Int [kHz] | 2.762E+2 | 1.891E+2 | 3.409E+2 | 1.258E+2 | 5.144E+1 | 1.016E+2 | 9.905E+1 | 4.313E+1 | ||||
| Int_corr [kHz] | 2.816E+2 | 1.920E+2 | 4.010E+2 | 1.408E+2 | 5.472E+1 | 1.085E+2 | 1.336E+2 | 6.187E+1 | ||||
| # samples | 735 | 735 | 619 | 619 | 213 | 213 | 66 | 66 | 30 | 30 | 19 | 19 |
| # cells | 295 |
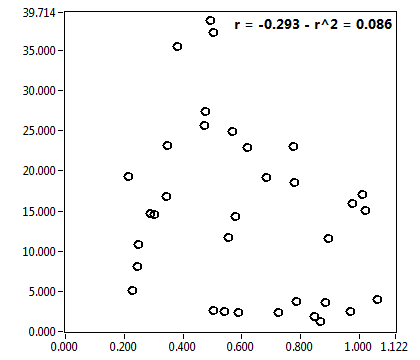
![c_corr [nM] - Ch1 - Nuc : ave = 3741.218 - sd = 2241.152](HP1hi_c_corr_1n.png)
![Int_corr [kHz] - Ch1 - Nuc : ave = 281.559 - sd = 191.957](HP1hi_Int_corr_1n.png)
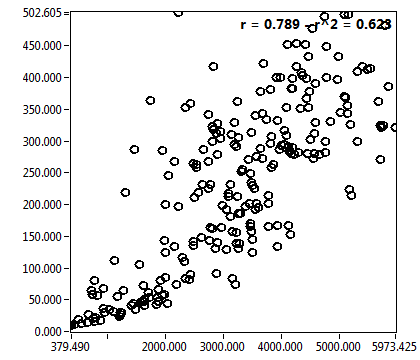
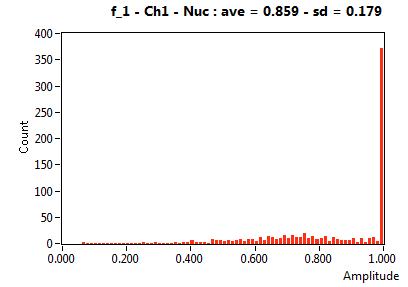
![D_1 [mum^2/s] - Ch1 - Nuc : ave = 2.470 - sd = 4.369](HP1hi_D_1_1n.png)
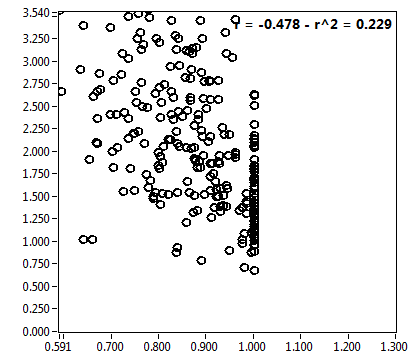
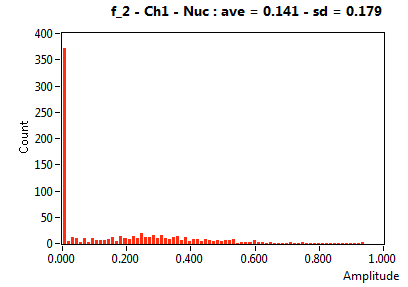
![D_2 [mum^2/s] - Ch1 - Nuc : ave = 0.217 - sd = 0.098](HP1hi_D_2_1n.png)
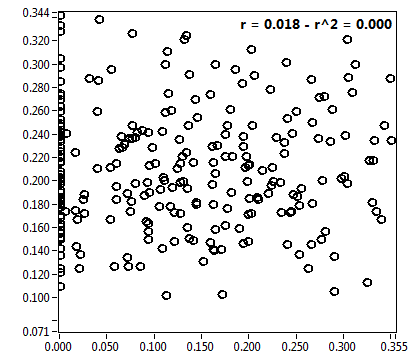
![CPM_corr [kHz] - Ch1 - Nuc : ave = 0.478 - sd = 0.255](HP1hi_CPM_corr_1n.png)
![D_m [mum^2/s] - Ch1 - Nuc : ave = 1.937 - sd = 2.887](HP1hi_D_m_1n.png)
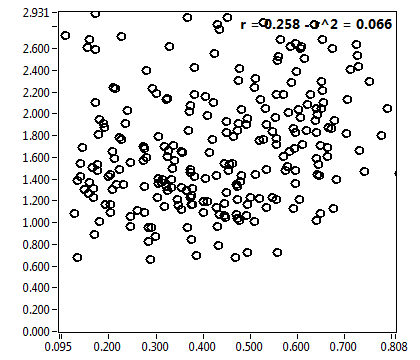
![c_corr [nM] - Ch1 - Nuc : ave = 3741.218 - sd = 2241.152](HP1hi_c_corr_1n.png)
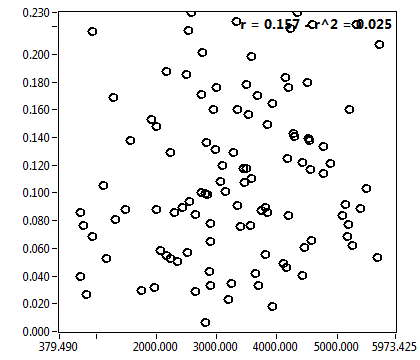
![c_corr [nM] - Ch1 - Cyto : ave = 425.587 - sd = 734.251](HP1hi_c_corr_1c.png)
![Int_corr [kHz] - Ch1 - Cyto : ave = 54.724 - sd = 108.469](HP1hi_Int_corr_1c.png)

![D_1 [mum^2/s] - Ch1 - Cyto : ave = 26.012 - sd = 24.872](HP1hi_D_1_1c.png)
![D_2 [mum^2/s] - Ch1 - Cyto : ave = 0.672 - sd = 0.428](HP1hi_D_2_1c.png)
![CPM_corr [kHz] - Ch1 - Cyto : ave = 0.709 - sd = 0.659](HP1hi_CPM_corr_1c.png)
![D_m [mum^2/s] - Ch1 - Cyto : ave = 23.931 - sd = 24.432](HP1hi_D_m_1c.png)